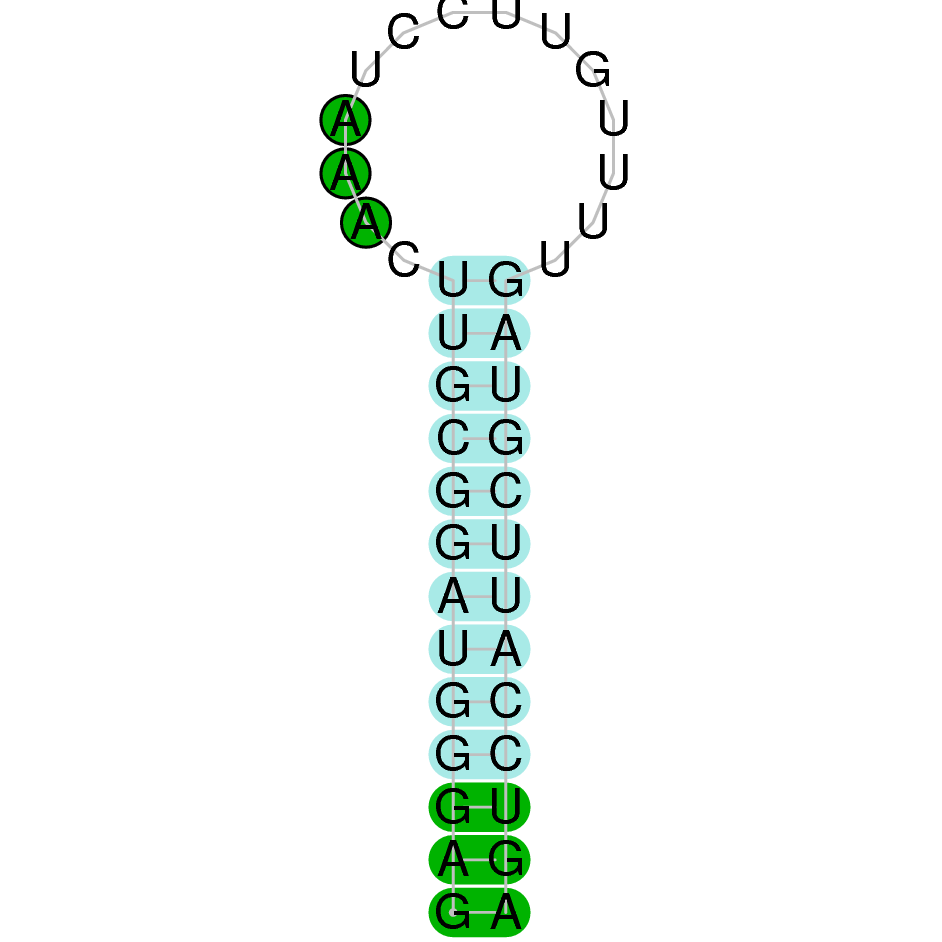Introduction
Selenoproteins
Selenium is an essential trace element in mammals due to its presence in proteins in the form of selenocystein (Sec) and its implication of the maintainance of important cellular and organismal functions(1,2). Selenocysteince Sec-containing proteins (selenoproteins) have been identified in all domains of life. The human selenoproteome is encoded in 25 selenoprotein genes(3). Sec is an amino acid structurally similar to cystein (Cys), differing in the presence of the selenium. Among others, Sec is the only known amino acid in eukaryotes whose biosynthesis occurs on its own tRNA, which is encoded by the UGA codon. This codon normal function is the translation stop, so organisms evolved the Sec insertion machinery to incorporate this amino acid at UGA codons, in order to avoid the stop of the translation. Cis-acting Sec insertion sequence (SECIS) are required to perform that(2). As said, selenoproteins mediate biological effects of selenium in the organisms, consequently, they are broadly present in the three domains of life: eukarya, archaea and eubacteria.
Selenoproteins biosynthesis

The biosynthesis of selenoproteins in either eukaryotes and archaea has been unknown for many years. Interestingly, Sec is the only aminoacid in eukaryotes whose biosynthesis occurs on its own tRNA (Sec tRNA [Ser]Sec). This tRNA has exclusive characteristics such as its length 90 nucleotides, which is much longest than normal tRNAs. It has also an incredibly long variable arm and very long acceptor stem and D-stem. Besides, both bacterial and human Sec tRNA [Ser]Sec present exceptionally secondary and tertiary interactions(2,3).
Biosynthesis steps:
- 1. Aminoacylation of tRNA[Ser]Sec:
- This step is conducted by the enzyme Seryl-tRNA synthetase (SerS). Thanks to different crystallographic studies it has been revealed that the interaction is produced between the variable stem of tRNA [Sec]sec and the NH2-terminal domain of SerS.
- 2. Seryl-tRNA[Ser]Sec phosphorylation:
- This is considered as an additional catalytic step that results on the synthesis of a O-phosphoseryl-tRNA[Ser]Sec intermediate substrate before the last step of the biosynthesis. It is executed by the action of the phosphoseryl-tRNA kinase (PSTK).
- 3. Last step:
- Finally the Sec synthase incorporates the active form of Se, which is selenophosphate, to the O-phosphoseryl-tRNA[Ser]Sec. This is how lastly the Sec-tRNA[Ser]Sec is synthesized.

Incorporation of Sec intro proteins is conducted by UGA codons that usually lead to the end of translation. In order to avoid this and promote the coding potential of UGA codons various trans-acting factors are required.
Secis elements are cis-acting stem-loop RNA structures located in the 3’-untranslated regions of eukaryotic mRNAs(2).
- SBP2: it contains three domains: an NH2-terminal domain which has a regulatory role; a Sec incorporation domain (SID) and a COOH-terminal RNA-binding domain (RBD) both required for SECIS binding activity of SBP2. SBP2 is a limitation factor for selenoproteins synthesis since its knockdown leads to a decreasement of selenoproteins expression and an overexpression of it enhance Sec incorporation.
- eEFSec: it has a “chalice-like” structure formed by four domains being the fourth one responsible of the interactions with SPB2. it is in charge of recruiting tRNA[Ser]Sec and insert Sec into a nascent protein chain in response to UGA codons. It stands out that it has a really high specificity for aminoacylated tRNA[Ser]Sec.
- L30: the majority of L30 proteins is associated with ribosomes but a small fraction is present in a ribosome-free form. It seems that it might be part of the basal Sec insertion machinery.
- eIF4a3: it acts as a regulatory protein contributing to the preferential translation of essential selenoproteins during Se deficiency. It makes this by masking the SBP2 binding site so it can prevent Sec incorporation.
- Nucleolin: it is a regulatory phosphoprotein involved in rRNA synthesis and ribosomal biogenesis. It stands outs its role regulating the hierarchy of selenoprotein expression.

Selenoproteins evolution in mammals
Selenoproteins are sec-containing proteins, which are encoded by UGA. Selenophosphate synthetases (SPS), the enzyme nedeed for its synthesis, is highly conserved in both eukaryotic and prokaryotic genomes that encode selenoproteins, being itself a selenoprotein in many species. Recent studies show that aquatic organisms generally have larger selenoproteomes than terrestrial organisms, and that mammalian selenoproteins show a trend toward reduced use of selenoproteins. The Marco Mariotti (3) study found 21 selenoproteins in all vertebrates: GPx1-4, TR1, TR3, DIO1, DIO2, DIO3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB1 , SelS, SelT1, SelW1 and Sep15. The other selenoproteins were found only in certain lineages, highlighting a dynamic process by which new selenoprotein genes were generated by duplication, while others were lost or replaced their Sec with cysteine (Cys). This study predict ancestral vertebrate selenoproteome (imatge selenoproteome.jpg), along with the details of tis transformations across vertebrates, specifically they found 28 proteins in the ancestral vertebrate selenoproteome and 25 in the ancestral mammalian selenoproteome(3).
Selenoprotein families
Glutation Peroxidases (GPx)
Gpx family are widespread in all three domains of life. In concrete, in mammals there are eight different GPx paralogs. Five of them contain a Sec residue in their active site (GPx1, GPx2, GPx3, GPx4 and GPx6) and the other three contain a Cys (GPx5, GPx7 and GPx8). This protein family is involved in hydrogen peroxide signaling, detoxification of hydroperoxides and maintaining cellular redox homeostasis. In particular GPx4 stands out from all the other GPx for many reasons. Firstly, GPx4 it is a monomeric protein that is expressed in a wide range of cell types and tissues. Its levels are less affected by Se availability than other GPx and it belongs to the housekeeping selenoproteins. There are three different alternatively spliced GPx4 mRNA isoforms that code for cytosolic (cGPx4), mitochondrial (mGPx4) and nuclear (nGPx4) proteins. The first one is expressed during embryonic development and in adult organs but mGPx4 and nGPx4 are only expressed in testes. In fact, it has been reported that the lack of mGPx4 causes male infertility in male mice, suggesting the role of GPx4 in role male gametogenesis. Referring to GPx4 physiological functions, this protein is involved in the reduction of complex phospholipid hydroperoxides and its specificity to them is due to the configuration of its active site, which lack a loop structure that allows access for bulky phospholipid hydroperoxide molecules. Besides this protective role by preventing lipid decomposition, GPx4 has been related to a redox-regulated cell death pathway that links to preventing neurodegeneration(2).
Thyroid Hormone Deodinases or Iodothyronine deodinases (DIO)
The iodothyronine deodinase family consist in three paralogous proteins in mammals (DIO1, DIO2 and DIO3), all of them involved in regulation of thyroid hormone activity by reductive deodination. In most vertebrates, the pro-hormone T4 has to be metabolized in order to obtain the T3 functional hormone. These three proteins have different localizations in cell and tissue expression: DIO1 and DIO3 are located on the plasma membrane, while DIO2 is localized to the endoplasmic reticulum(ER)(2).
- DIO1 is in charge of the metabolization by deiodination of T4 into T3 hormone in pheriperal tissues such as liver and kidney.
- DIO2 can also performe the deiodination of T4 into T3 and is essential for the maintaining of T3 levels in the brain during the critial period of development.
- DIO3 can only metabolize T4 into triiodothyronine (rT3), which is biologically inactive. It has an important role in the regulation of thyroid hormone inactivation during embryological development.
Thioredoxin reductases (TXNRDs)
Thioredoxin reductases (TXNRD) family are oxidoreductases that, together with thioredoxin (Trx), compromise the major disulfide reduction system of the cell. Specifically, in mammalian cells, there are three different TXNRD isozymes (TXNRD1,TXNRD2 and TXNRD3) that contain a Sec residue in the COOH-terminal penultimate position . These TXNRD are flovoenzymes that contain selenocysteines and have a major role in reducing thioredoxins, as well as in redox homeostasis. The three isozymes differ, among others, in the cell localization and its function:
- TXNRD1 is mainly localized in the cytosol and nucleus. There are at least six different TXNRD isoforms in mammals, and each one is involved in different processes. For instance, the isoform 1 possess glutaredoxin activity as well as thioredoxin reductase activity, moreover, it is implicated in the induction of actin and tubulin polymerization, leading to formation of cell membrane protusions. In contrast, among other functions, isoforms 4 and enhances the transcriptional activity of estrogen receptors.
- TXNRD2 is localized in the mitochondria. This one mantains thioredoxin in a reduced state and it is implicated in the responses against oxidative stress. It may play a role in redox-regulated cell signalling as well.
- TXNRD3, as the first one, this one displays also glutaredoxin and glutathione reductase activities. In addition, TXNRD3 plays an important role in sperm maduration as it promote the formation of sperm structural components.Specifically, it catalyzes disulfide bond isomerization and promotes disulfide bond formation between GPx4 and sperm proteins (2,3).
Selenoprotein R or Methionine-R-Sulfoxide Reductase 1
Methionine-R-Sulfoxide Reductase 1(MSRB1) is a zinc-containing selenoprotein that was previously identified as Selenoprotein R and Selenoprotein X. MSRB selenoproteins function is the stereospecific reduction of methionine-R-sulfoxide to methionine. This methionine oxidation is also a post-translational modification acting as a regulator of actine assembly, which is ultimately promoting filament repolymerization. In mammals, MSRB1 is the main MSRB protein and it is localized in the cytosol and in the nucleus. However, there are two additional homologs (MSRB2 and MSRB3) that show lower levels of expressions in mammals with a similar catalytic efficiency to MSRB1. Moreover, their localization is also different, MSRB2 is located in mitochondrias, whereas MSRB3 is located in the ER (2).
Methionine sulfoxide reductases A (MsrA)
MsrA is a highly conserved selenoprotein and it catalyzes the enzymatic reduction of both free methionine-S-sulfoxide and its protein-based form by using thioredoxin. The MSRB family, explained above, was so named because of its functional similarity to MrsA. Despite they are structurally different they have complementary functions(2).
15-kDa selenoprotein (Sel15)
Also known as Sel15, this selenoprotein is a thioredoxin-like fold ER-resident protein which has a distinct Cys-rich domain in the NH2-terminal domain. This is necessary to interact with UGGT, which is an ER-resident chaperone involved in a protein quality control in the ER. I has been shown that Sel15 expression is induced by conditions related to an accumulation of misfolded proteins in the ER (6). Moreover its expression is regulated by dietary Se and also belongs to the stress-related selenoproteins. In fact, studies suggest the possible role of Sel15 regulating the chemopreventive effects of dietary(2).
Selenoprotein I
This selenoprotein is only found in vertebrates. SelI is a transmembrane protein compound by seven transmembrane domains and three conserved aspartic residues which are key for the catalytic function(2).
Selenoprotein M
SelM is located in the perinuclear region and it is highly expressed in the brain. It has been suggested its role as a neuroprotective protein. In fact, in humans it has been linked to familial early onset Alzheimer’s disease and hepatocellular carcinoma(2).
Selenoprotein N
SelN was one of the first selenoproteins identified thanks to bioinformatic. It is a transmembrane glycoprotein highly expressed during embryonic development and this make sense with the fact that mutations in SelN gene (SEPN1) have been related to muscle disorders known as SEPN1-related myopathies. Different studies with zebrafish and SelN knockout mice suggest that this selenoprotein has an important role in the maintenance of satellite cells and that is needed for the regeneration of skeletal muscle tissue after stress or injury. Moreover, it has been suggested that SelN acts as a cofactor of the ryanodine receptor (Ryr), which mediates the release of calcium from the sarcoplasmic reticulum during muscle contraction. Therefore SelN may be involved in the regulation of intracellular calcium transport(2).
Selenoprotein O
This selO was discover more than a decade ago and even nowadays it have not been characterized any of its structural or biochemical features. It contains just one Sec residue which is located in the antepenultimate position at the COOH-terminal end. SelO’s function still remain unknown, as well as the function as its homologs which have been detected in many species such as bacteria or plants, for example (2).Selenoproteins P
SelP represents almost the 50% of the total Se in plasma and its unique feature is the presence of multiple Sec residues. It is mainly synthesized in the liver although its mRNA is expressed in all tissues. ApoER2 and megalin are two receptors that have been shown to be involved in the uptake of SelP in brain and testis. Regarding the relation between these receptors and SelP, studies suggest the role of SelP in transporting Se to peripheral tissues, in particular brain and testis (2).
Selenoproteins K and S
These two selenoproteins can be considered as one family due to their topology and it is one of the most widespread eukaryotic family They consist of a single transmembrane domain in the NH2-terminal sequence, a glycine rich segment and a characteristic location of Sec residues in the COOH-terminal end of the proteins. However, SelS differs from SelK because of the presence of and additional coiled-coil domain in the cytosolic portion of the protein. They are located in the ER membrane and belong to the type III group of transmembrane proteins. These two selenoproteins have been recently implicated in ER-associated degradation (ERAD) of misfolded proteins. In fact, it has been seen that they interact with each other through their transmembrane domains apart from through additional proteins. Referring with their role in ERAD, it is known that their genes contain functional ER stress response elements and that the expression os both proteins is up-regulated by the conditions that promote the accumulation of misfolded proteins in the ER (2,3). Besides the role of SelK and SelS in ERAD these two proteins might be involved in the regulation of anti-inflammatory effects of Se and the immune system(6).
Selenoproteins H, T, V and W
These selenoproteins belong to the Rdx family of selenoproteins, which are thiol-based oxidoreductases. They are characterized for having a thioredoxin-like fold, a conserved Cys-x-x-Sec motif and a conserved stretch of amino acids in the COOH-terminal portion of the protein with the tGxFEI(V) consensus sequence (2).
SelH
This selenopretin was firstly identified in fruit flies as BthD protein and then they found its homologs in the human genome. SelH includes a Sec residue within the Cys-x-x-Sec motif and a conserved nuclear targeting RKRK motif in the N-terminal sequence. SelH has a unique subcellular localization pattern which was found specifically in the nucleoli. This selenoprotein is sensitive to dietary Se intake. besides, it has been seen that SelH specifically binds to sequences that contain heat shock and stress response elements and that has glutathione peroxidase activity (2).
SelT
It is predominantly localized to the ER and Golgi and it is ubiquitously expressed during embryonic development but also in adult tissues. It has been suggested that SelT has an important role in the regulation of calcium homeostasis and neuroendocrine function. Moreover recent studies report its implication in the regulation of pancreatic beta-cell function and glucose homeostasis (2).
SelW
It is one of the most abundant selenoproteins in mammals. It is located in the cytosol and it is highly expressed in muscles and brain. Moreover it is highly regulated by the availability of Se in the diet. Besides that, it has been reported that SelW could be involved in redox regulation of 14.3.3 protein which is the ubiquitous signaling adapter protein. Two more different paralogs of SelW exists apart from SELW1, which are SELW2 and SELW3 (2).
SelV
This selenoprotein may has evolved by duplication from SelW. It is only found in placental mammals, nevertheless it has been specifically lost in some of them such as gorillas. It is larger than SelW because of the presence of an additional NH2-terminal domain. About its physiological role, it has been seen that SelV is only express in testes so it may be involved in male reproduction (2).
Selenoproteins U
SelU was firstly found in fish. In the case of humans there are three subfamilies: SelU1, SelU2 and SelU3. They belong to the thioredoxin-like superfamily and they have redox activity (2).
Selenoprotein J
SelJ stands out because it has a very restricted phylogenetic distribution, in fact it does not exist in mammalian genomes. It appears to be restricted to actinopterygian fishes and sea urchin and shows significant similarity to the jellyfish J1-crystallins. Moreover, in contrast to the rest of the selenoproteins it does not appear to have any function role but structural (2).
Selenoprotein L
SelL belongs to the thioredoxin superfamily. As well as the SelJ it has a restricted phylogenetic distribution, in concrete, in aquatic organisms. Besides, it is not found in mammals(2).
Selenoprotein E
SelE is also called Fep15 due to its relation to the Sel15. This protein is localized in the ER and its function is yet unknown. It is not relevant for out research as it is only found in fish(2).
Mungos mungo
Mungos mungo commonly known as banded mongoose is a mongoose species native from Southern Africa. It is mainly distributed south of the Sahara, but this range extends across Africa, from Gambia to north-eastern Ethiopia and down to South Africa. (7)
Taxonomy
| Eukaryota | |
| Animalia | |
| Chordata | |
| Mammalian | |
| Carnivora | |
| Herpestidae | |
| Mungos | |
| Mungos mungo |
Physical description

The banded mongoose is a small one, as adults reach approximately 30 to 45cm of length and a weight of 1.5 to 2.25 kg. It has a large head, small ears, short and muscular limbs and a long tail. A curious characteristic is that animals of wetter areas are larger and darker coloured than animals of dyer regions. They are characterised by a greyish brown and dark rough fur with several dark brown to black horizontal bars across the back. These bands make these mongooses distinguishable from other species. The limbs and snout are darker than the rest of the body. The colour of the nose varies from brown to orange-red.(7,8)
Habitat and ecology
The banded mongooses have a broad habitat tolerance. For instance, they inhabit in savannahs, open forests, grasslands, woodlands and rocky country. They also live near water as well as in dry. However, the banded mongoose is not found neither in desert or semi-desert areas(7). The mongooses use usually various types of dens for shelter, such as termite mounds. Nevertheless, they also live in rock shelters, thickets, gullies and warrens under bushes, due to its capacity for living in such different habitats(7,8).
Social behaviour
This animal is diurnal and gregarious, living in packs from 10 to 20 members, in constant change. The pack usually stays together in the same area as a group for living, but they leave the group individually to forage for food. As a pack, these mongooses are nomadic and they will not stay in one particular den or sheltering for too long, in fact, they usually move every 2-3 days. In some situations, like when no refuge is available or they are being attacked by predators, the group will form a compact arrangement in which they lie on each other with heads facing outwards and upwards, as a self-defensive mechanism (7,8,9).

Generally, the aggression between them is low and there is no strict hierarchy in mongoose groups. Most aggression behaviour occurs during oestrus. Sometimes they may squabble for food, but typically the one who claims the food first wins. When groups are too large, some females are forced out of the group by either older females or males. These displaced females usually form new groups with subordinate males. (7) In terms of reproduction, females become sexually mature around 9 to 10 months of age, and unlike other mongoose species, all females of the group can breed. Females participate actively in courtship and males reciprocate. Gestation is about 2 months (60-70 days) and litter size is variable, ranging from two to six pups. Characteristically, reproduction within a pack is often synchronized so that several females give birth in the same week. During the first weeks of life, mongoose pups stay in the dens where they are guarded by a single helper, generally a young nonbreeding male or breeding female. By the time of three months of age they become nutritionally independent (8,9).
Conservation
There are protected areas in Africa where banded mongooses live. The abundance is variable depending on the area, for instance the Serengeti of Tanzania has a density around 3 mongooses/km2 while the Queen Elizabeth National Park in Ugana has much bigger density, 18 mongooses/km2(10). It is not a threatened species, since its conservation state is Least Concern according to the IUCN Red List (10).
MATHERIALS AND METHODS
The aim of our study was to identify and annotate the selenoproteome of Mungos mungo, thus all the selenoproteins and the machinery required for their synthesis. In order to do that, Homo sapiens selenoproteome was selected as the reference genome, both because it has a very well annotated selenoproteome and because its known selenoproteins are very conserved, especially within mammals. The next figure shows an overall scheme of the program structure:
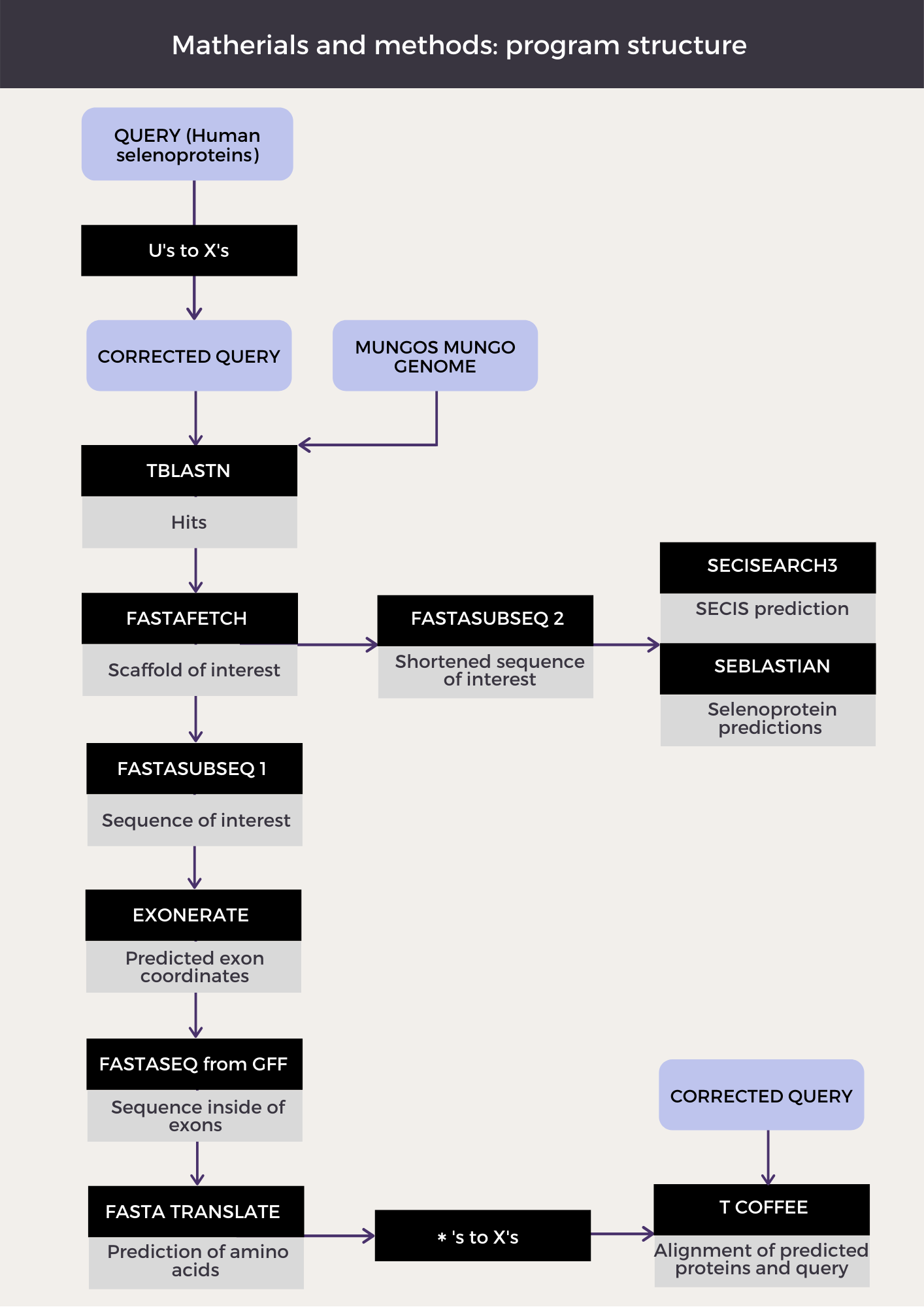
Obtention of the genome and the index:
The Mungos mungo genome was downloaded from the following source:
/mnt/NFS_UPF/soft/genomes/2019/Mungos_mungo/genome.fa
The indexed genome was obtained from:
/mnt/NFS_UPF/soft/genomes/2019/Mungos_mungo/genome.index
QUERY ACQUISITION
As previously mentioned, the Homo sapiens genome was used to identify the selenoproteome of Mungos mungo. We used the database SelenoDB 1.0 (http://www.selenodb.org//) to check and obtain the amino acid sequences of the different selenoproteins found in Homo sapiens. Every sequence was copied into a gedit file named ‘protein’.fa, where ‘protein’ is the abbreviation of each selenoprotein name as found on SelenoDB 1.0. For instance: Sel15.fa stands for the 15-kDa selenoprotein.
All of the proteins were downloaded and stored in a directory named proteins. In addition, all the symbols not corresponding to an amino acid, such as # or $ found between amino acids or at the end of the sequence were removed.
Since our aim was to predict selenoprotein sequences encoded in the Mungos mungo genome, we queried those human selenoproteins in our mongoose genome through the use of different algorithms, as we will now explain.
PROGRAM
We developed a Perl program in order to automate the most part of genome annotation. The code for the program is the following: Program.
In the first place we created a loop in order to be able to analyze all of the query proteins found in our directory. We designed this loop so every time a new protein was run, a new folder -named as the protein- would appear in the directory. All of the program outputs and intermediate documents would get automatically saved in this directory. We also added a second loop in case we wanted to run a protein more than once.
So as to ease the analysis process, we decided to name our output files in an orderly way. The name of all of our outputs starts with the name of the protein; in case a certain scaffold has been selected, its name appears then and also does the number of iterations that the program has done for that protein; lastly, the name of the program is written in the output file. The purpose of adding the iteration number to the name of the outputs is to help the program from overwriting protein predictions when two hits from the same query are being analyzed. An example to that would be:
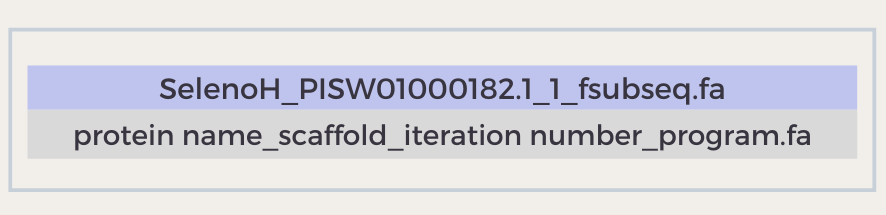
The output document seen above would result from running the program FastaSubSeq for the protein SelenoH, having selected the scaffold PISW01000182.1; the number 1 between the names of the scaffold and the program indicates that this is the first time this protein is run with our program.
Before starting running the different proteins, since some programs show recognition issues when the software encounters a “U” character (selenocysteine) we implemented an algorithm so the U’s found in our queries are replaced to X’s.
The analyzed selenoproteins were the followings (34):
With the aim to make a thorough analysis of the Mungos mungo selenoproteinome, we used a series of different programs:
TBLASTN
tBLASTn is a Blast version that compares protein sequences against a nucleotide sequence database translated in all six reading frames. In our case, we are querying our human selenoproteins against the mongoose genome. Thus, this program outputs a series of hits -regions were proteins match, and thus, are likely homologous to our query- and orders them by e-value and scaffold. Since we are only interested in good e-values (very close to 0) we added a threshold of 0.001 to our outputted hits.
FASTAFETCH
FastaFetch is a program designed to extract our scaffolds of interest, which are the ones that we got from the best tBLASTn hits.
FASTASUBSEQ
The next step to our proteome annotation is FASTAsubSeq. It extracts the chromosomal region were your hits are found. The presence of introns makes it necessary to select a wide region of interest, ideally of about 200.000 base pairs. Issues appear when selecting the initial position and the length of this region. The problem is that not all of the scaffolds can fit regions that long, so this is solved this way:
We define the start location of our hit
my $start=$hit_start-'100000';if ($start>='0') { $start=$start; }else {$start='0'}Where “$hit_start” is a manually entered number obtained from the tBLASTn results and “$start” will be defined as the start position of our region of interest.
[...]#We check if the whole length we are selecting fits inside of our scaffold
if ($length_scaffold > ($start+'200000')){my $length='200000';} else {$length=($length_scaffold-$start);}Where “$length_scaffold” is a value obtained from:
/mnt/NFS_UPF/soft/genomes/2019/Mungos_mungo/genome.lengths and “$lenght” will be defined as the length of our region of interest.
This step is done twice, since in order to run Seblastian and SECISearch3, the region of interest can not be greater than 100.000 base pairs.EXONERATE
Exonerate is an algorithm that enables us to know how many exons can be found in the sequence we are entering and provides its relative coordinates.
FASTASEQFROMGFF
Once the exon coordinates are obtained, FastaSeqFromGFF extracts the coding base pairs, excising the intronic sequences. Thus, it obtains the sequence that encodes our target protein (cDNA).
FASTATRANSLATE
In this step, what we do is to translate the predicted cDNA to a protein, which ideally should resemble a lot the human selenoprotein, to which it is an homolog. At this point, since this program generates outputs that contain “*” symbols where selenocysteine should be found, all of the “*” were substituted by X’s.
TCOFFEE
TCOFFEE is actually a validation step. This program aligns the predicted mongoose selenoprotein to its original query, the human selenoprotein and provides a score to it. TCOFFEE generates several trash documents in the directory the program is found. Those can be deleted since the proper alignment is found in the directory that contains the rest of the outputs.
SECISEARCH3 AND SEBLASTIAN
This step is also a validation of the obtained results. It was run manually for each selenoprotein, since this two programs are found on-line. SECISearch3 is an algorithm that predicts feasible SECIS structures found in the chromosomal region of interest where our selenoprotein is supposed to be. SEBLASTIAN is a method for selenoprotein gene detection that uses SECISearch3 and then predicts selenoprotein sequences encoded upstream of SECIS elements. An ideally well-predicted mongoose protein should have viable SECIS (in the same strand of the protein and in the 3’ end) and match a known selenoprotein. Still, it has to be considered that SECISearch3 and SEBLASTIAN are highly automated algorithms, thus, they are less sensitive than manual classification systems.
PHYLOGENETIC TREE
In order to ensure that the predicted proteins within families were correct, phylogenetic trees have been built. For this purpose, the website Phylogeny.fr was used. This tool allows the comparison of multiple molecular sequences to analyze and establish their phylogenetic relationship. The input given to the website was generated creating a multifasta file containing the human proteins and the predicted Mungos mungo proteins for each family analyzed.
In order to run our program, the next commands should be executed:
→ Downloading the program and saving it at some directory, like “Documents” or “Desktop”
→ Downloading the human query selenoproteins, which can be found in the folder “proteins”
$ cd proteins/$ pwd
/home/uXXXXXX/location/proteins$ cd /home/uXXXXXX/program_location/$ chmod u+x PROGRAM.pl$ ./PROGRAM.pl /home/uXXXXXX/location/proteinsRESULTS
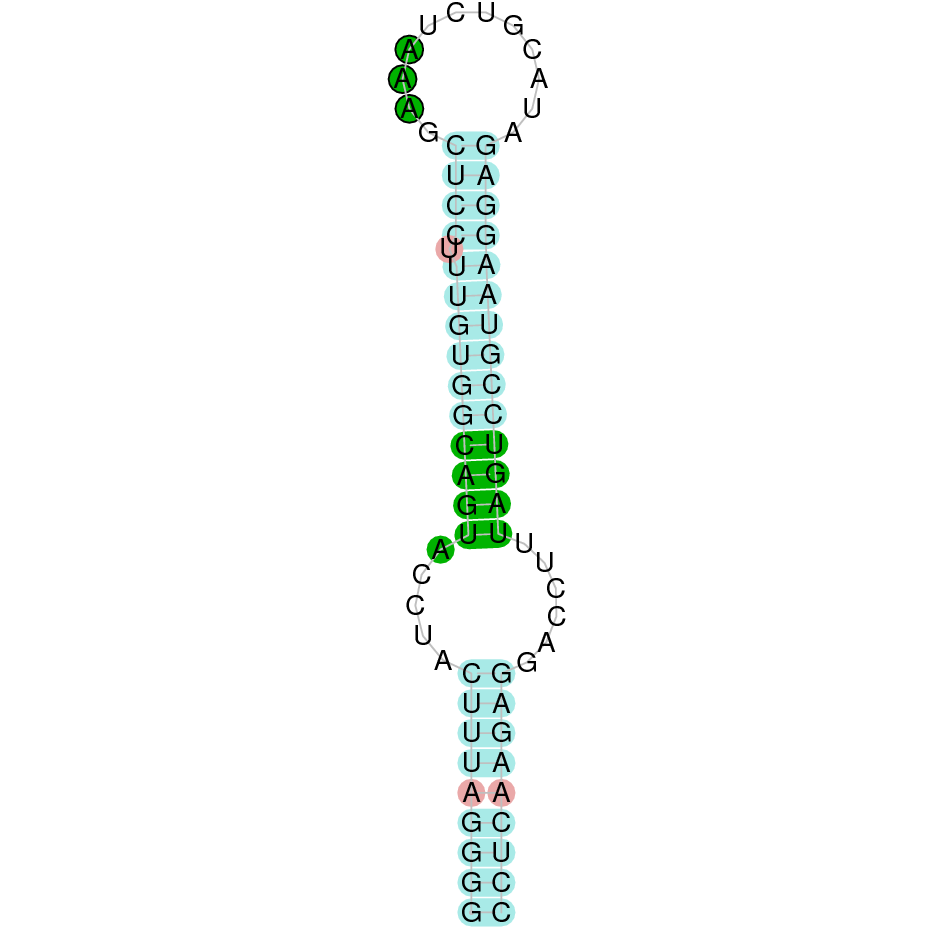
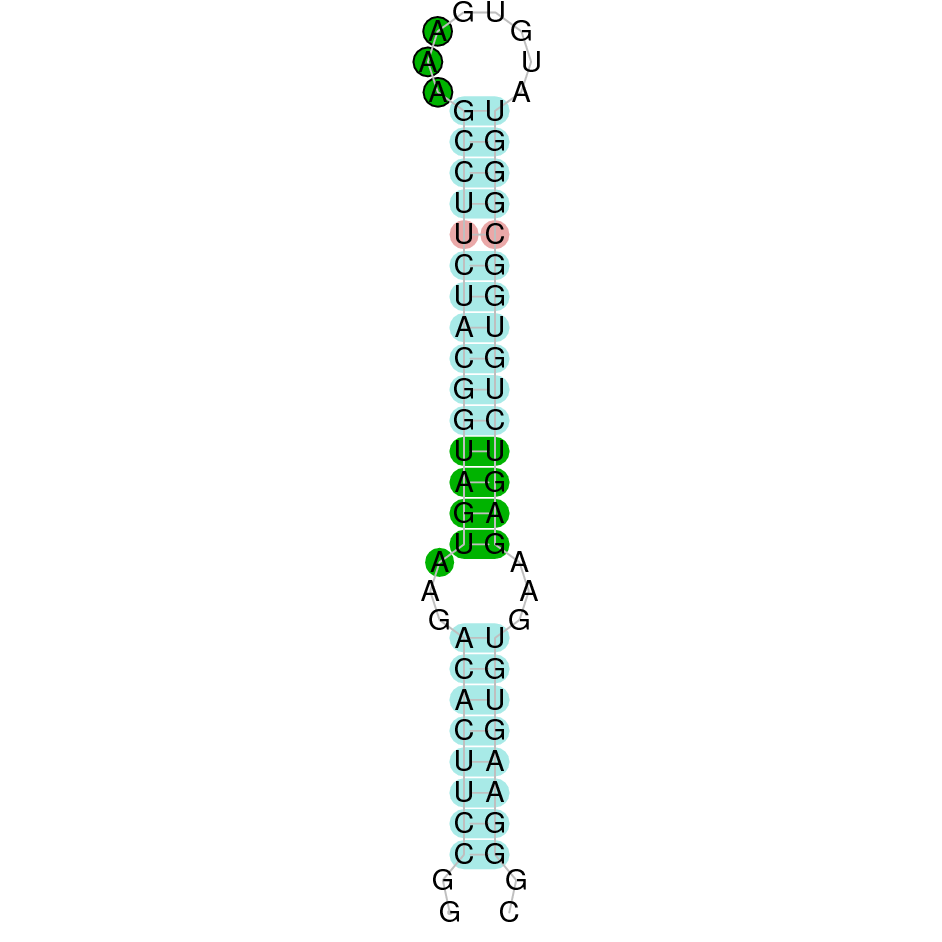
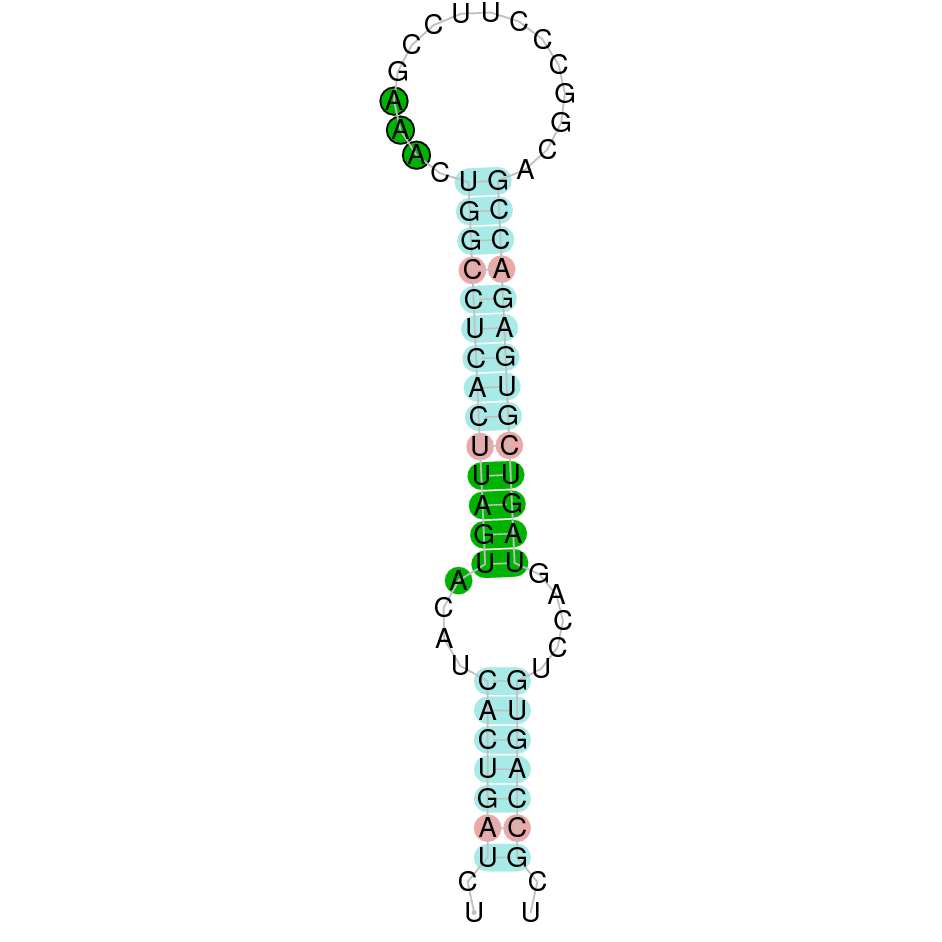
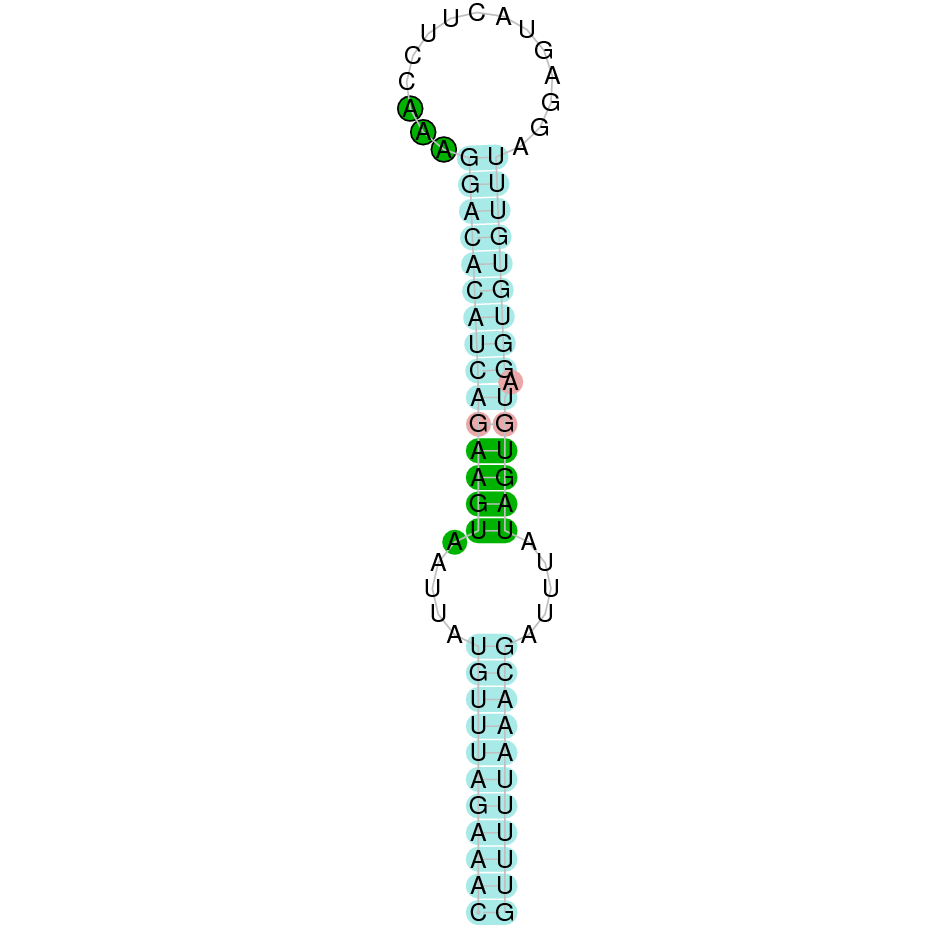
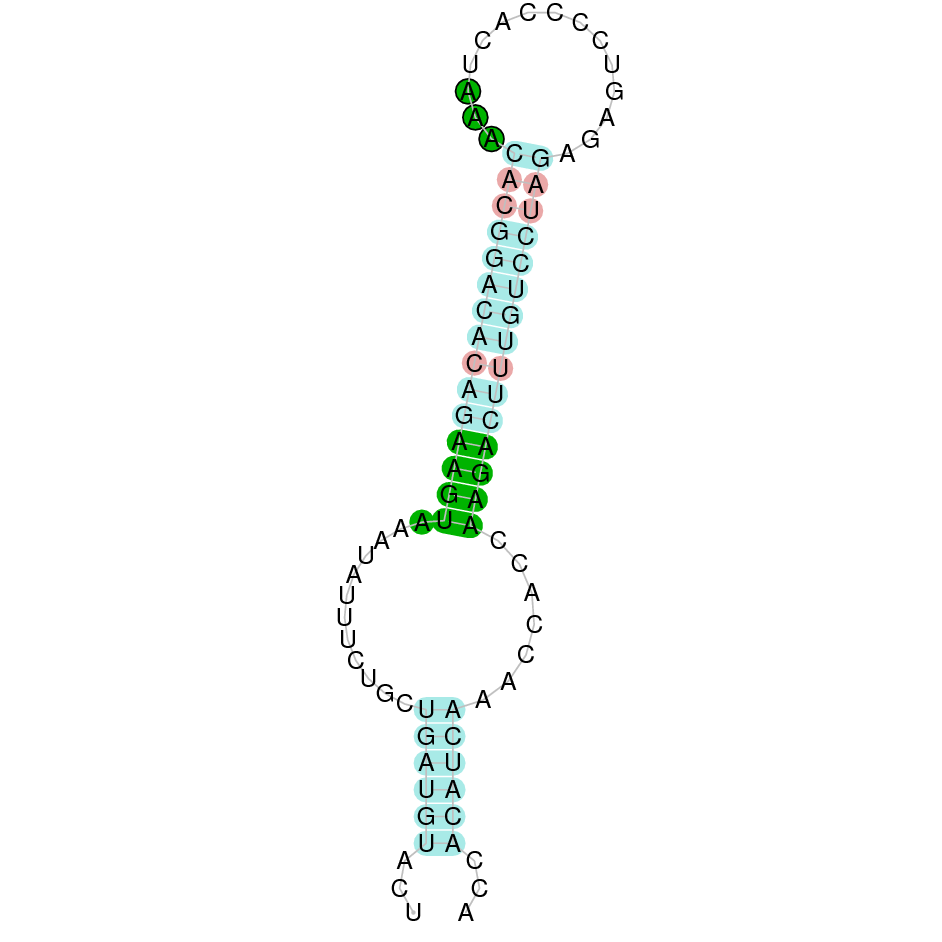
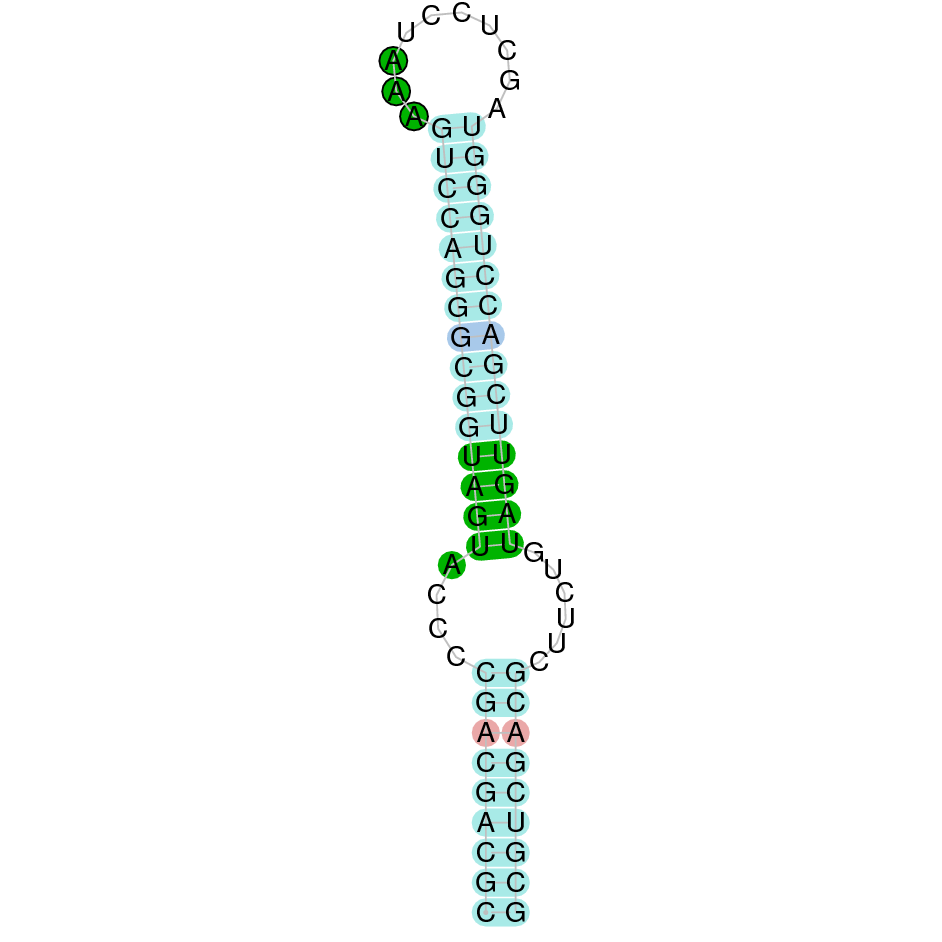
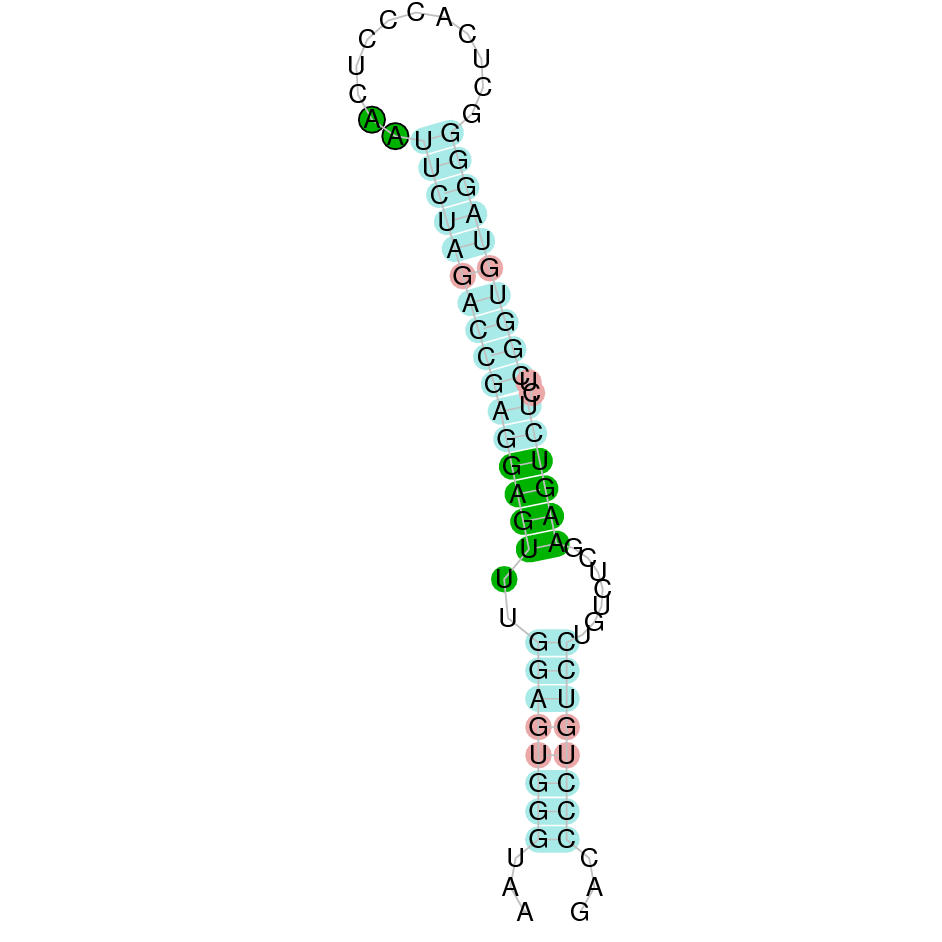
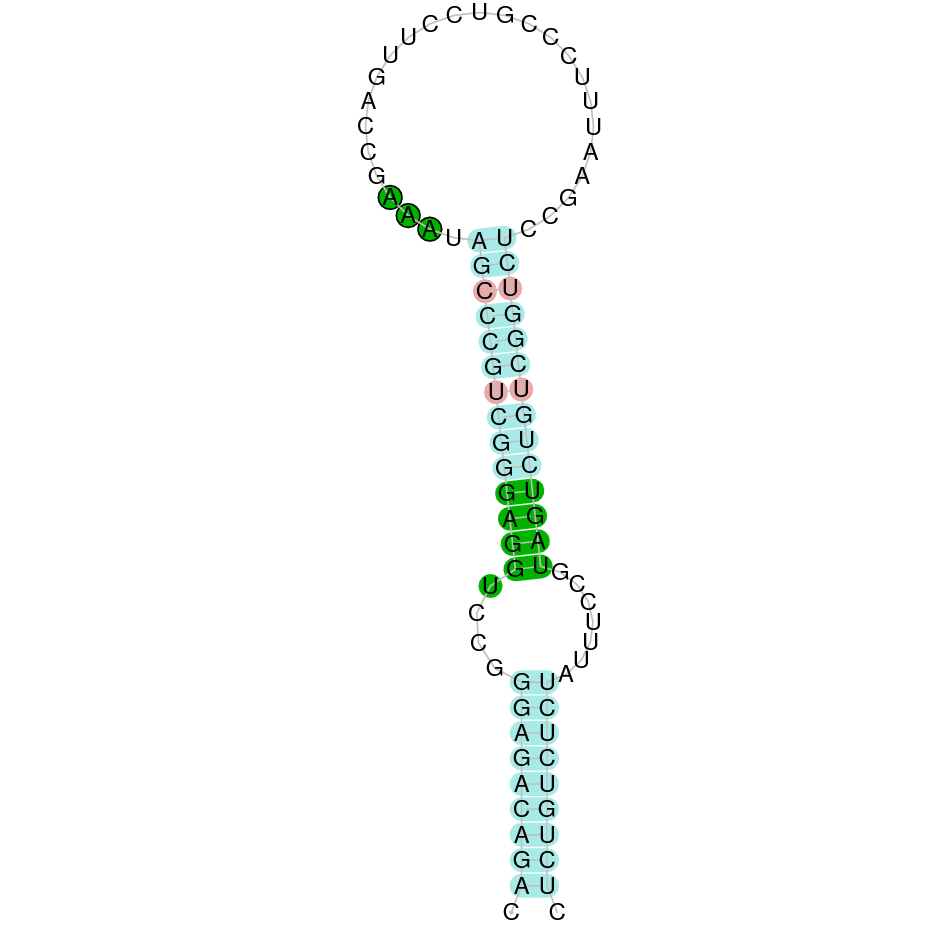
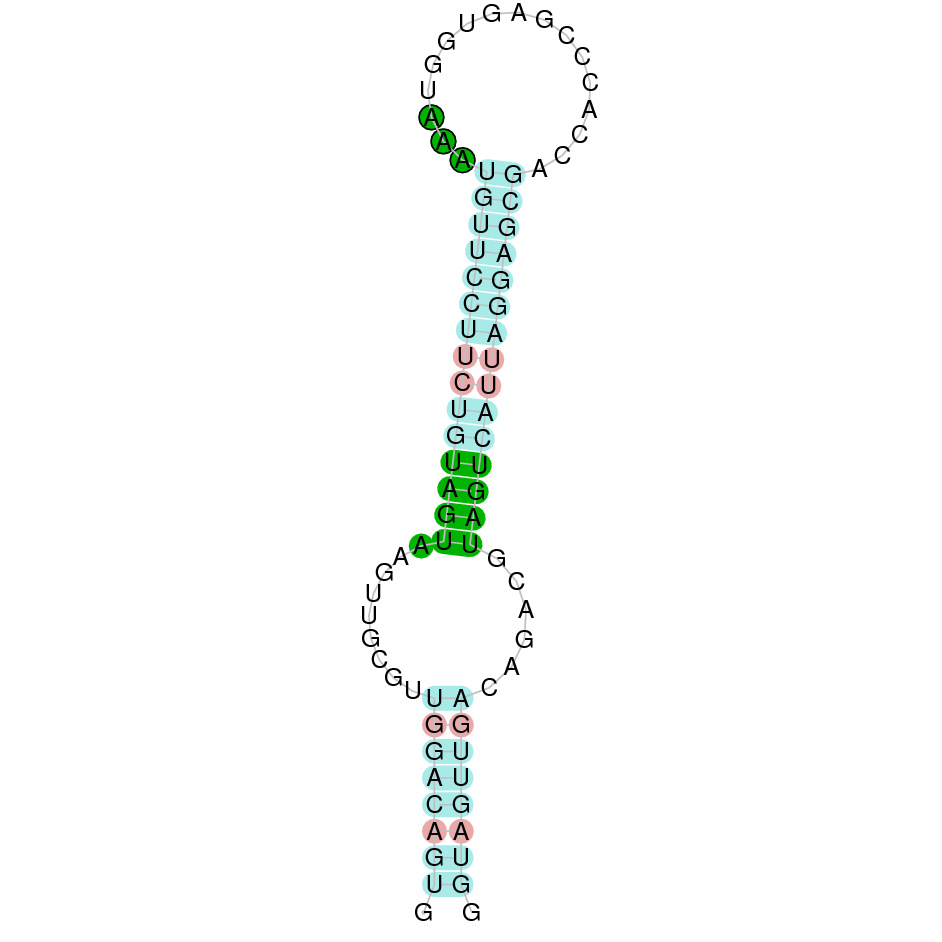
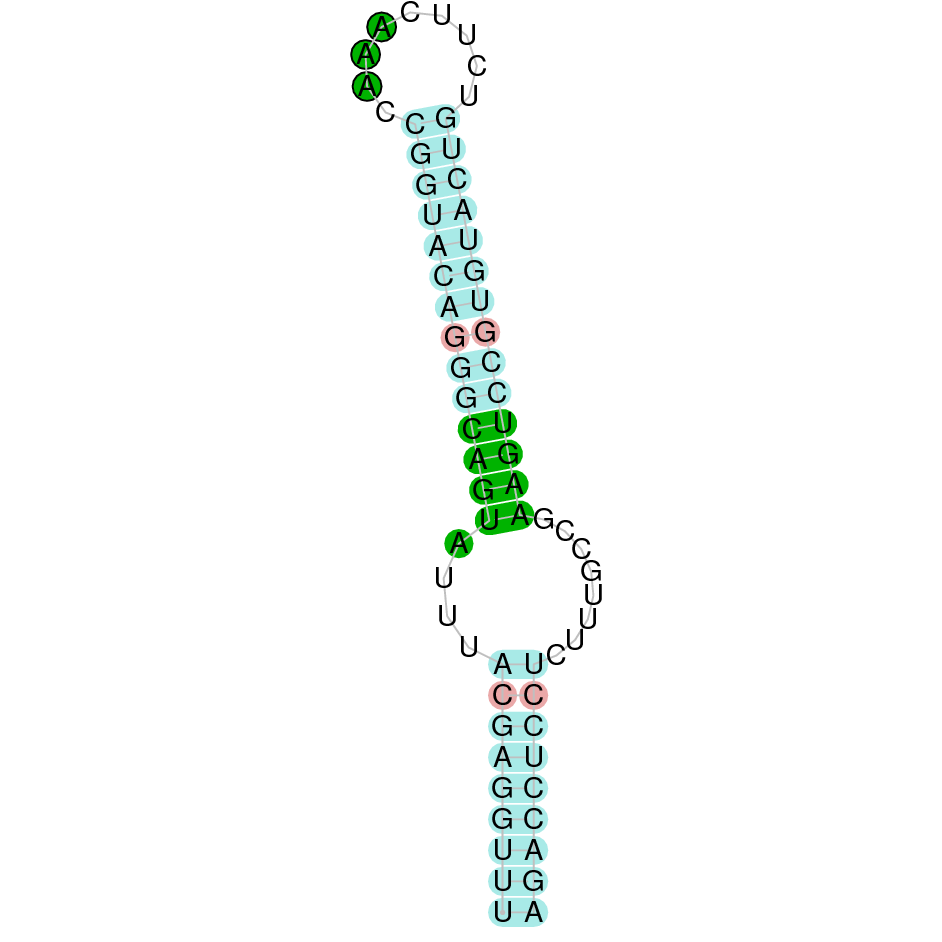
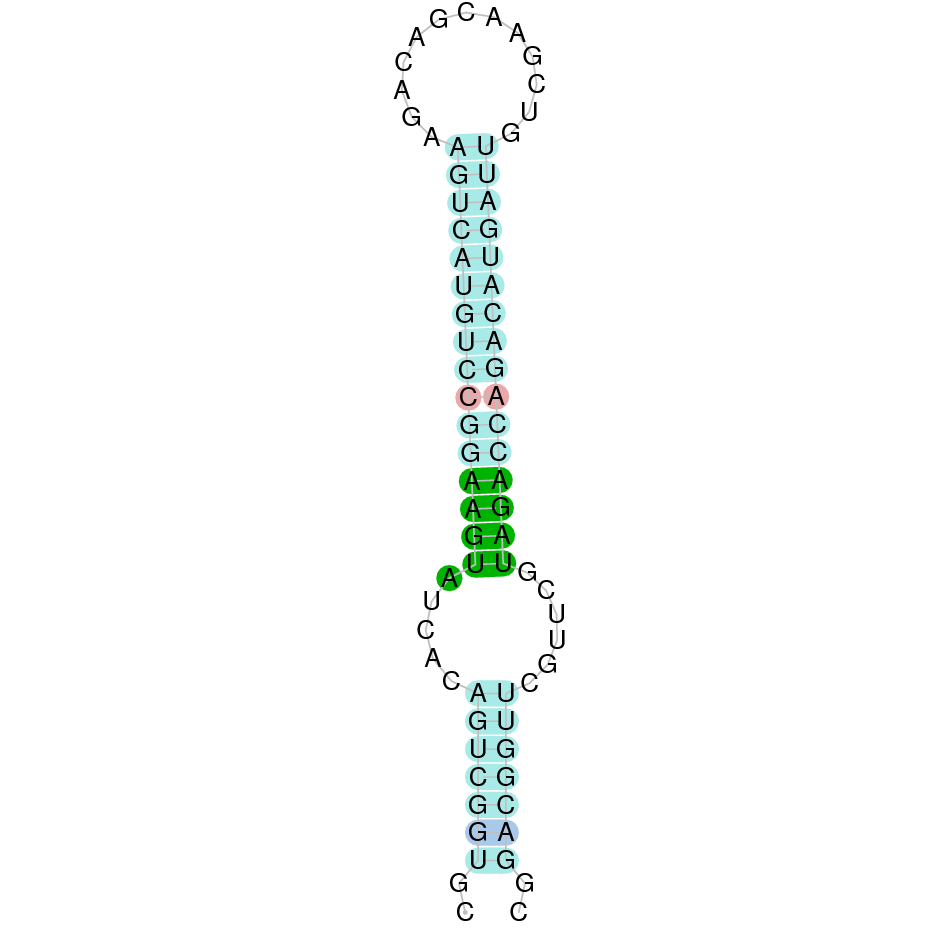
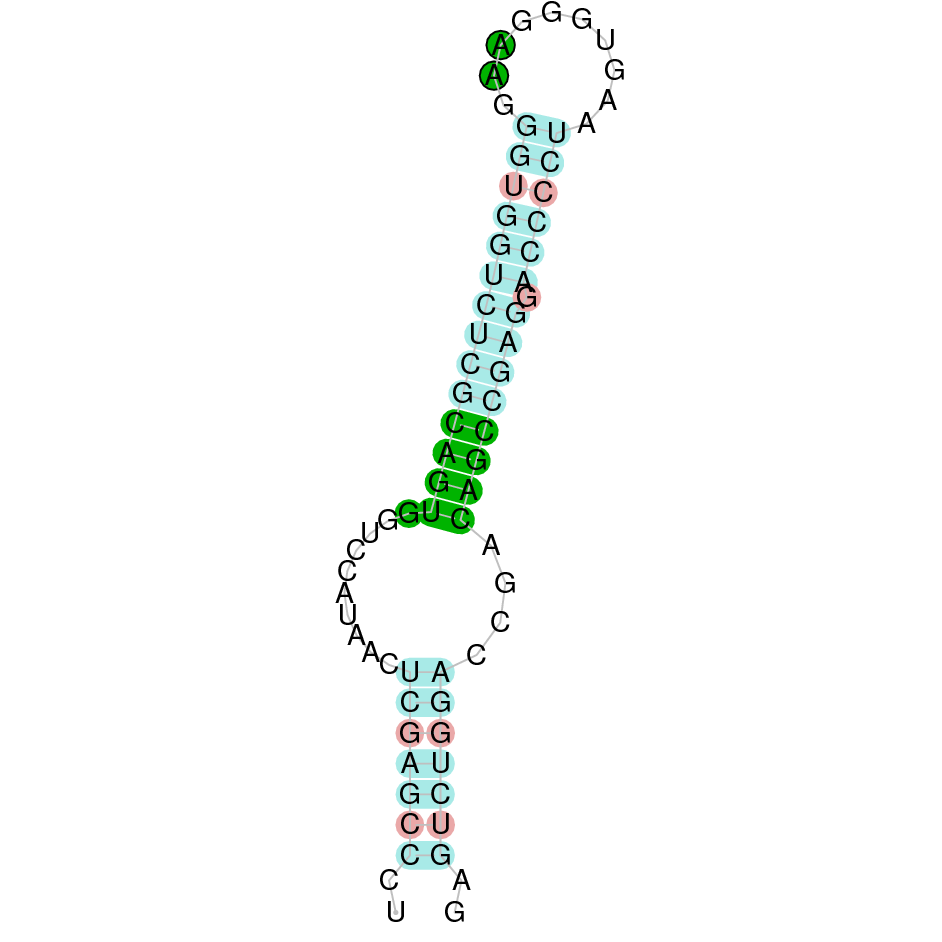
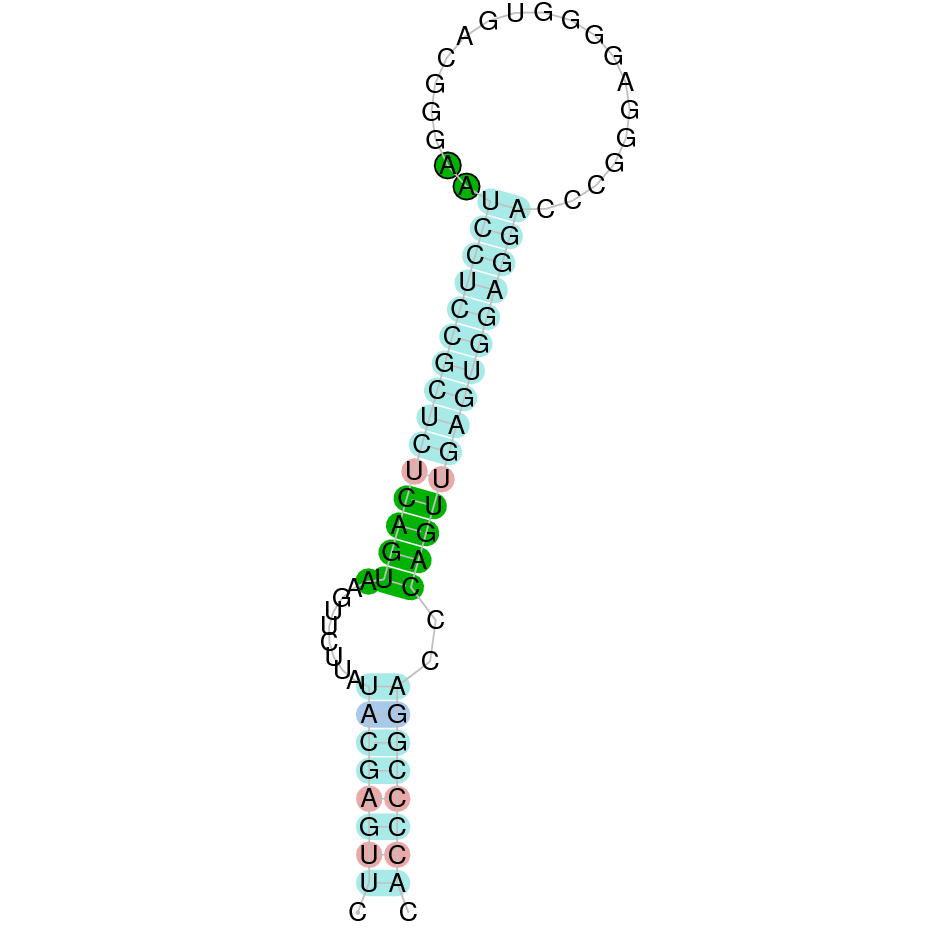
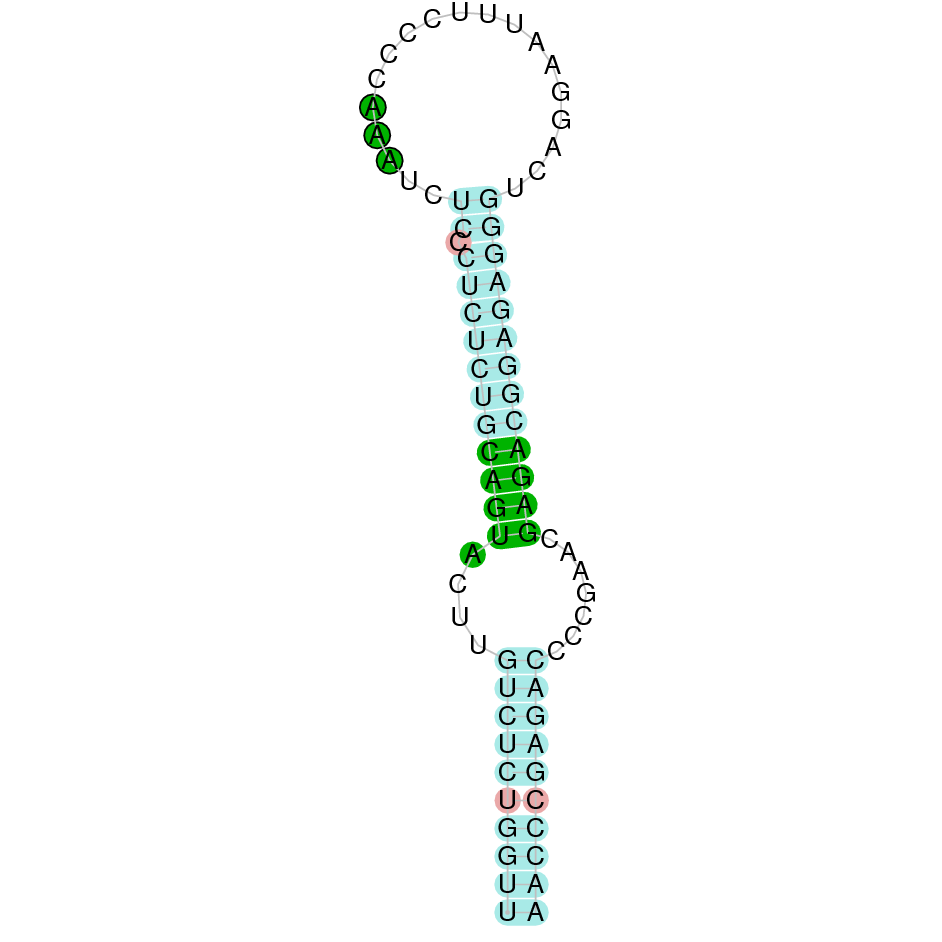
Here is shown the table results for the selenoproteins prediction in the genome of Mungos mungo. Specifically, it is shown for each query of the genome reference(Homo sapiens), the Tblastn output, the scaffold selected for the prediction, the localization of the gene and wether is a forward or a reverse strand, the exonerate prediction and the T-coffe alignment. Moreover it is shown the SECIS element prediction by Seblastian and SECISearch3, the SECIS resume prediction, the SECIS element image and finally, our predicted protein sequence. The legend used in the table is the following:

DISCUSSION
Selenoproteins
Glutation Peroxidases (GPx)
As said in the introduction, glutathione preoxidases are the largest selenoprotein family in vertebrates and are also widespread in all three domains of life. Specifically in mammals, there are eight GPx paralogs, from all of them, five contain a Sec residue in their active site(GPx1,GPx2,GPx3,GPx4 and GPx6) while in the other three GPx homologs (GPx5,GPx7 and GPx8) the Sec residue is replaced by Cys, being Cys-containing homolgs. It should be underlined that in some mammals those are not selenoproteins and hace a Cys in the active site instead a Sec.
The discussion of these proteins should be done looking all at once because its high grade of homology. Specifically, it is known that there are three evolution groups: GPX1/Gpx2, GPx3/GPx5/GPx6 and GPx4/GPx7/GPx8. Thus, it was necessary to analyse every blast results with a clooser look to avoid overlapping between the scaffolds. Regarding that,
while Blast was being evaluated we saw that in all proteins appeared the same hits in the same scaffolds and all their e-values were good. So, to finally assign each scaffold to its correct protein we took into consideration other features such as the hit lenght to compare it with the query lenght obtained by SelenoDB, or the T-coffee score and its characteristics, or the identity score. To do that, we have studied more than one scaffold for every protein to finally decide the better prediction taking into account all the previous aspects.
In Mungos mungo, we found that GPx1, GPx2 and GPx3 are selenoproteins while GPx5, GPx6, GPx7 and GPx8 are Cys-containing homolgs. Regarding the GPx4, it was impossible to determine its nature, since the residue align with the human Sec was missing due to a not-well-annotated genome of this species.
In order to better understand the evolution of this protein family between the two species we were comparing, we elaborated a phylogenetic tree:
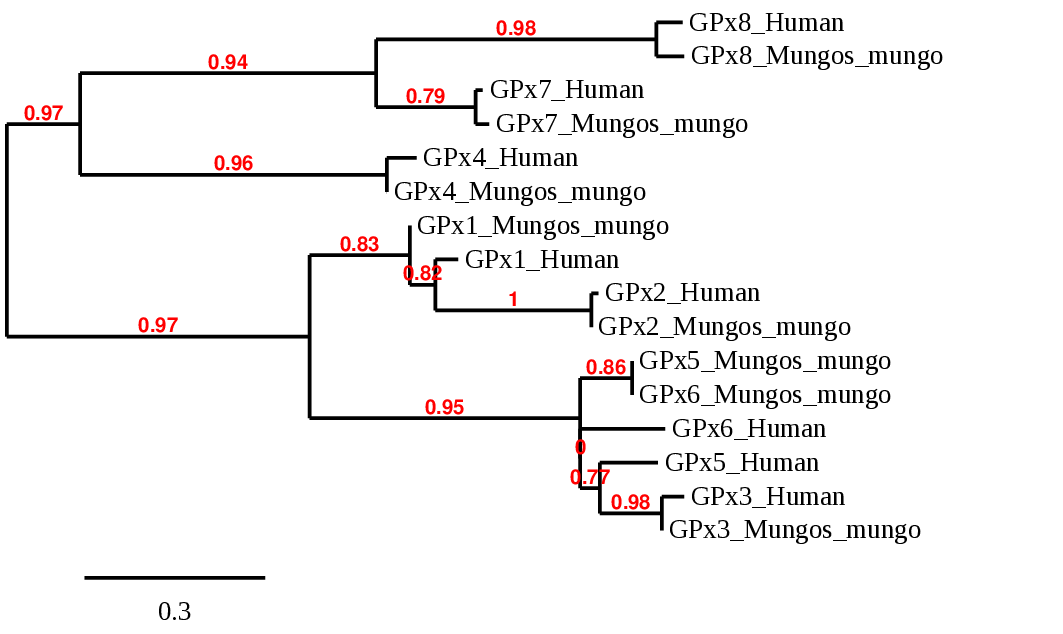
Regarding the phylogenetic tree, we can see that the GPx1, GPx2, GPx3, GPx4, GPx7 and GP8 Mungos mungo predictes proteins pair specifically with their homolog in humans. With the exception of GPx5 and GPx6, which was difficult to discriminate between the scaffolds obtained, so the predicton is not as accurate as expected and could led to misinterpretations. Even so, we can see that is confirmed the fact that GPx5 and GPx6 come from a GPx3 duplication since they are quite related. Furthermore, the tree confirm also the relation between GPx1, GPx2 and GPx3/5/6. By the other hand, we can also see that GPx7 and GPx8 are related to GPx4, indicating that those evolved from GPx4. In conclusion, the phylogenetic tree obtained confirm that our predicted selenoproteins are homologs of the human ones, and they evolved in the same way.
GPx1
The gene that encodes the protein GPx1 is located in the scaffold PISW01001800.1 between the positions 318681-319483 in the reverse strand. Among all possible scaffolds obtained on the Tblastn, we selected that scaffold since it was the one with the best and highest e-value (1.01e-68) and its 2 alignment fragments has an average identity of 92,675. The gene has 2 exons predicted with exonerate,located at positions 319271-319513 and 318681-319034, both in the negative strand. The T-coffee shown an excellent score of 991.Moreover, it contains a Sec in the same position as human GPx1 protein does and the predicted protein has almost the same lenght as the human one. However, the predicted protein doesn’t start with a Methionine as human does, it starts with an Alanine (A). With all that, we can afirm that Mungos mungo has the GPx1 selenoprotein. Seblastian was able to predict a grade A SECIS element between positions 318558-318627 at the 3’ end of the negative strand as well as the GPx1 selenoprotein in this sequence.

GPx2
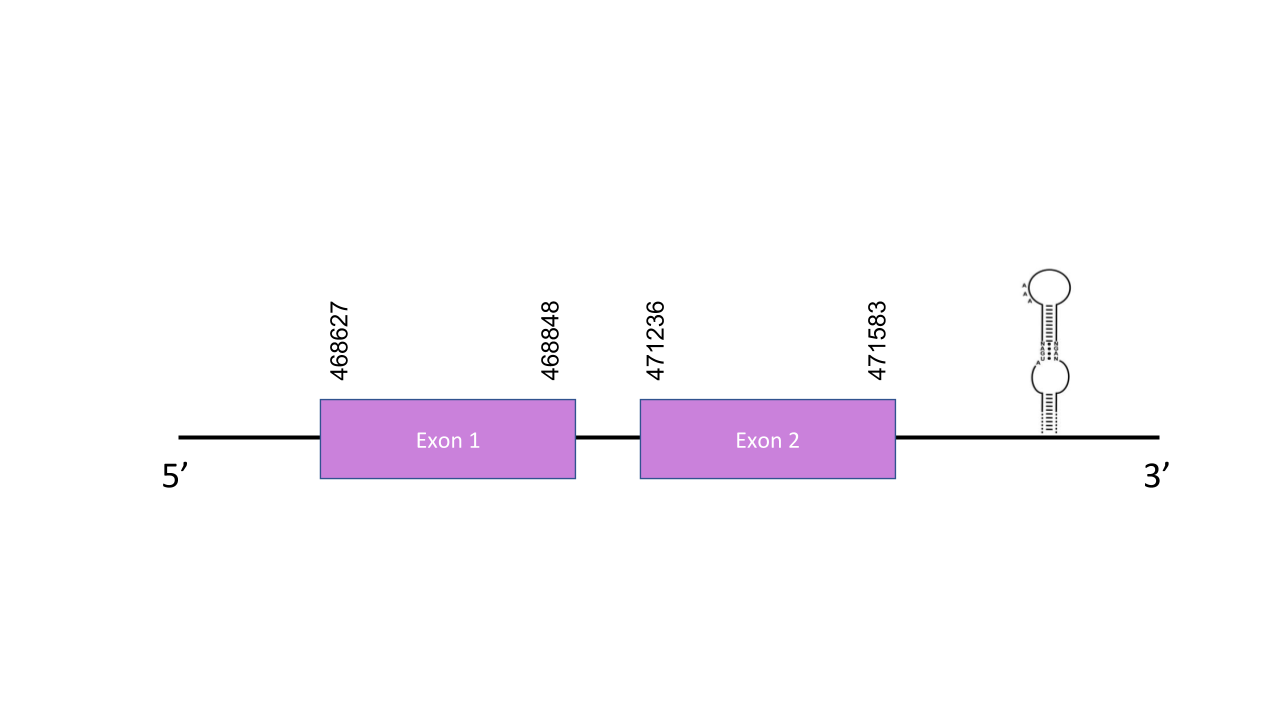
The gene that encodes the protein GPx2 is located in the scaffold PISW01000412.1 between the positions 468627-471583 in the forward strand. Among the 12 scaffolds obtained on the Tblastn, we selected that scaffold since it was the one with the best and highest e-value (2.96e-71) and its 2 alignment fragments has an average identity of 95,05. This gene has 2 exons predicted with exonerate,whose coordinates were 468627-468848 and 471236-471583, both in the positive strand. The T-coffee shown an score of 1000, which indicates a perfect alignment between the two species sequences.Moreover, the predicted protein starts with a Methionine and has a Sec residue, confirming that the predicted GPx2 is a selenoprotein in Mungos mungo. In this sequence, Seblastian shown two SECIS elements, both with an A grade, the first one located between 471808-471871 and the second one between 493771-493844, both at the 3’ end on the positive strand.However, just the first one was considerated valid, since the second one is more than 200.000 positions away the end of our final exon. It predicted the GPx2 selenoprotein in this sequence as well.
GPx3
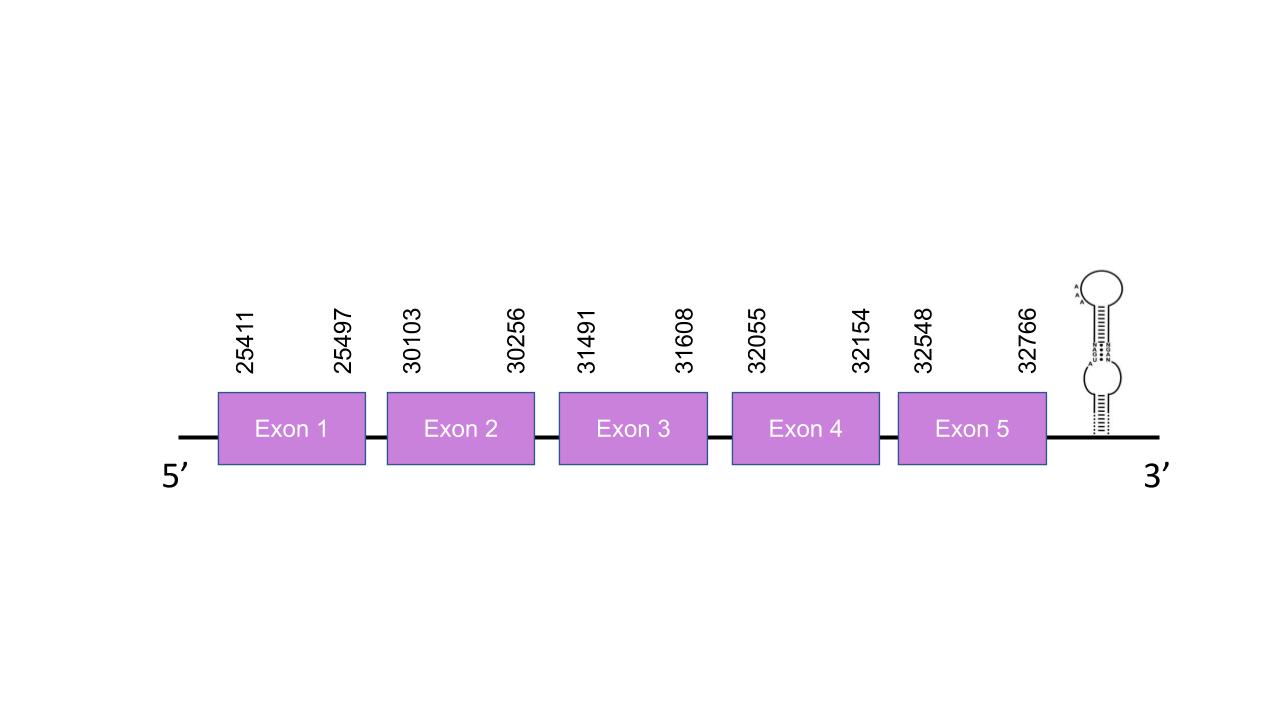
The gene that encodes the protein GPx2 is located in the scaffold PISW01005197.1 between the positions 30100-32766 in the forward strand. Among all the scaffolds obtained on the Tblastn, we selected that scaffold since it was the one with the best and highest e-value (6.21e-37) and its 4 alignment fragments had an average identity of 89,50. This gene has 5 exons predicted with exonerate,located at 25411-25497, 30103- 30256, 31491-31608,32055-32154 and 32548-32766, all of them in the positive strand. The T-coffee shown an score of 1000, which indicates a perfect alignment between the two species sequences.Moreover, the predicted protein starts with a Methionine and has a Sec residue, confirming that the predicted GPx3 is a selenoprotein in Mungos mungo. In this sequence, Seblastian was able to identify two SECIS elements, the first one was a grade A SECIS between 33383-33456 on the positive strand, while the second one was a grade B SECIS between 31374-31455 positions on the positive strand too. This last one was considered unvalid since it was in the middle of the predicted exons. Otherwise, the first SECIS found was in the 3’ end so this is a viable structure since is located in the same strand of our predicted gene and after it. The GPx3 selenoprotein was predicted correctly as well.
GPx4
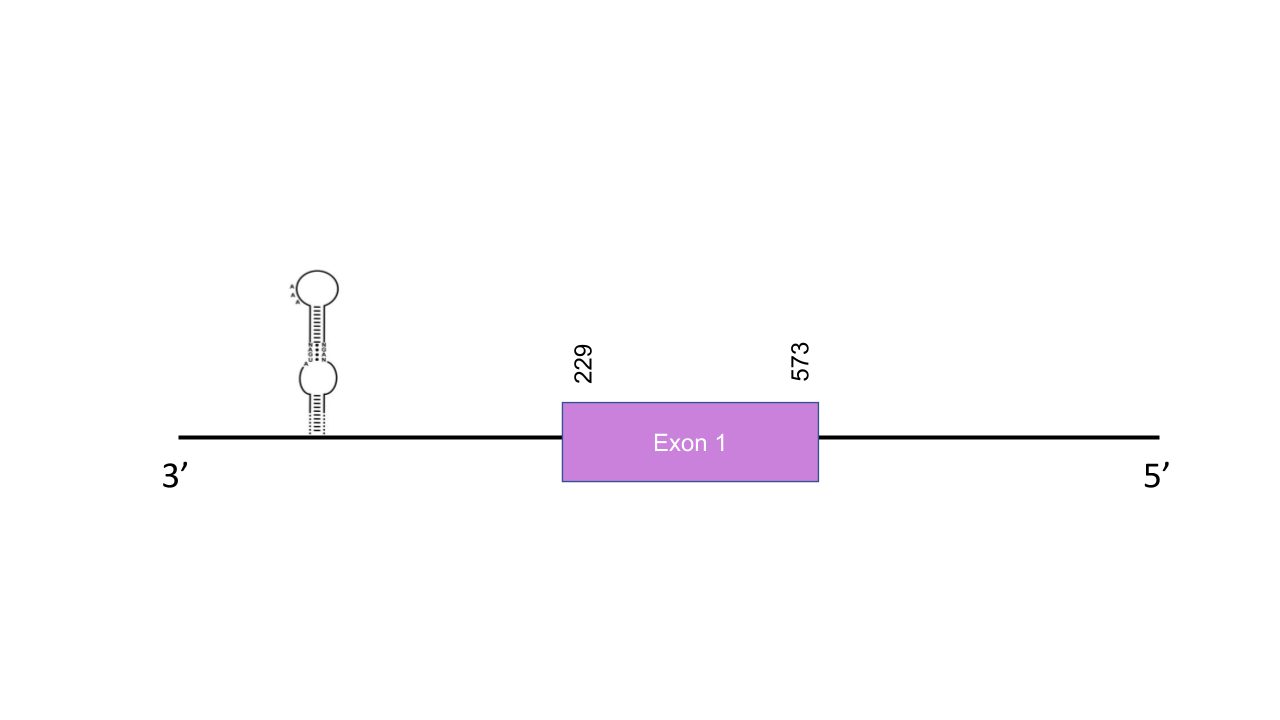
The gene that encodes the protein GPx4 is located in the scaffold PISW01043757.1 between the positions 229-573 in the reverse strand. We selected that scaffold since it was the one with the best and highest e-value (6.21e-37) and has an identity of 93.103. Exonerate shown a single exon located at 229-573 positions of the negative strand. T-coffee shown an score of 999, which describes a very good alignment. However, there was a huge residue-gap at the beginning of the sequence, and precisely, a gap in the residue where human protein shown a Sec residue. Thus, probably this part of the genome was not correctly annotated. Knowing that, we cannot confirm whether this residue corresponds to a Sec or a Cys. Thus, the selenoprotein GPx4 cannot be predicted correctly. In contrast, Seblastian was able to found a grade A SECIS element between 96-168 positions, so it is a valid structure since it is located in the same strand of our predicted gene and after it on the 3’ end. This may indicates that there was a Sec residue in the gap position alineated with the human Sec, but we are not able to confirm that since it is not well-annotated.
GPx5 and GPx6
As said before, these two proteins have evolved together from a GPx3 duplication. Keeping that in mind, as we expected, GPx5 and GPx6 share the same scaffold hits in the Tblastn, located at the negative and positive strand, respectively. They also share similar positions with the scaffold selected for GPx3 as shown hereunder.
GPx5
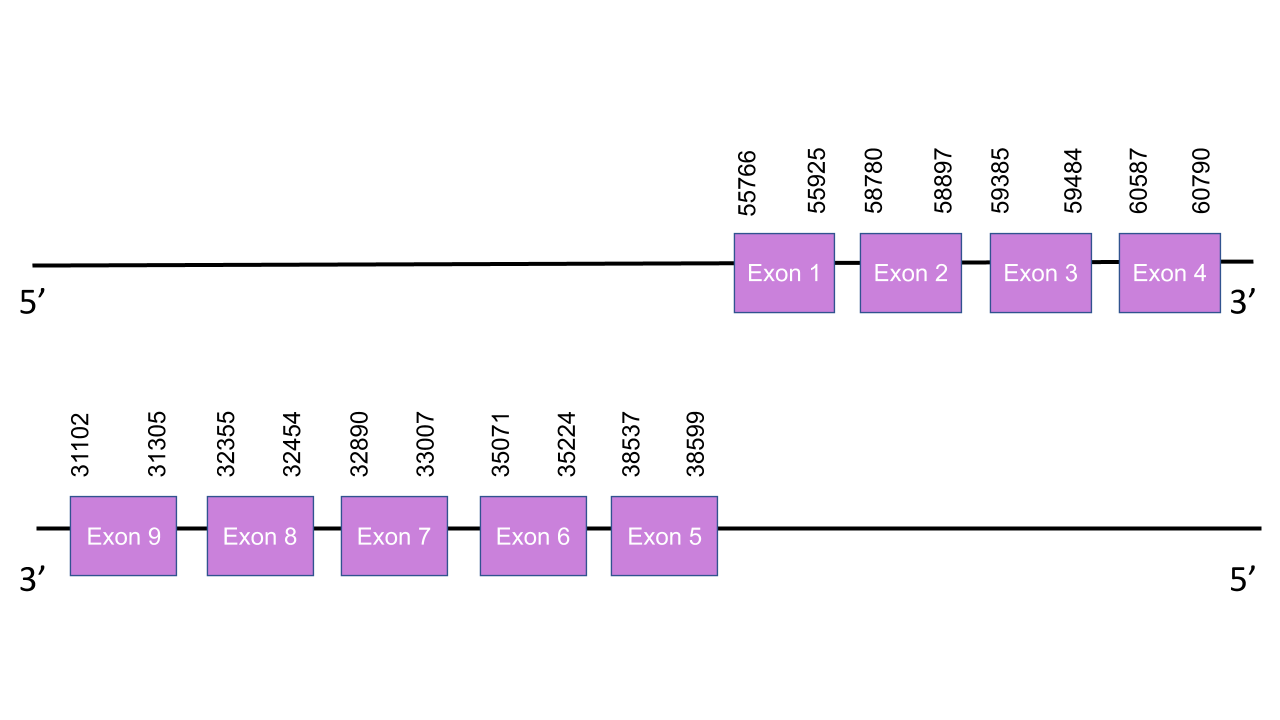
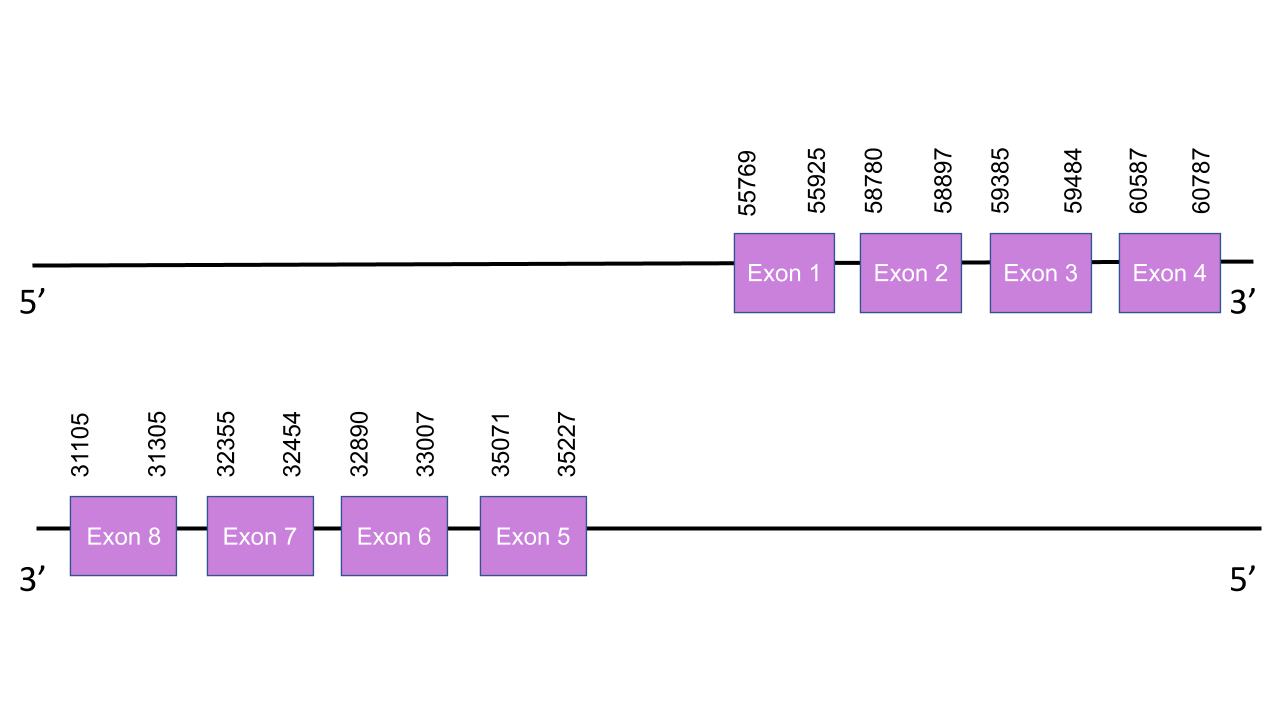
The gene that encodes the protein GPx5 is located in the scaffold PISW01008701.1 between the positions 31102-60790 on the reverse strand.Among the 10 scaffolds shown by Tblastn, this was the selected one,after an exhaustive study, due to its characteristics as it was the one with the lowest e-value and the higher average of identy in all its 8 alignment fragments. Indeed, it was taken into account the T-coffee results to take this decision. The gene presents 9 exons in the following absolute positions: 55766-55925(+), 58780-58897(+), 59385-59484(+) and 60587-60790(+) and 38537-38599(-), 35071-35224(-), 32890-33007(-), 32355-32454(-), 31102-31305(-). That means the four first exons are in the positive strand while the last five exons are in the negative strand. The t-coffee shown an score of 983, so is a very good aligment between the two species. Although our sequence doesn’t start with a Methionine, it has many cysteine well conserved, which indicates, as expected, that GPx5 is a Cys-containing homolog. Seblastian was unable to predict any SECIS element nor protein, which makes sense with the fact that our protein only presents cysteines, meaning that it might present a loss of selenoprotein function.
GPx6
The gene that encodes the protein GPx6 is located in the scaffold PISW01008701.1 between the positions 31105-60787 on the forward strand.Among all the scaffolds shown by Tblastn, this was the selected one due to its characteristics as it was the one with the lowest e-value and the higher average of identy in all its 8 alignment fragments, same as happens with the GPx5 analysis. This gene has 8 exons in the following absolute positions: 55769-55925(+), 58780-58897(+), 59385-59484(+) and 60587-60787(+) and 35071-35227(-), 32890-33007(-), 32355-32454(-), 31105-31305(-). That means the four first exons are in the positive strand while the other four exons are in the negative strand. 55769-55925(+),58780-58897(+),59385-59484(+),60587-60787(+),35071 35227(-),32890-33007(-),32355-32454(-),31105-31305(-). Same as happens in GPx5 the four first exons are located in the positive strand while the last five are in th negative one. The t-coffee shown an score of 970,which is a very good aligment between the two species. However, our predicted protein starts with a Glycine (G) instead of a Methionine, as human protein does. It is important to underline that we expected this protein to be a selenoprotein, with a Sec residue, but in Mungos mungo the Sec residue has been replaced by a Cys residue. Thus, GPx6 is a Cys-containing homolog in this animal. Seblastian was unable to predict any SECIS element nor protein, meaning that this protein might present a loss of selenoprotein function.
GPx7
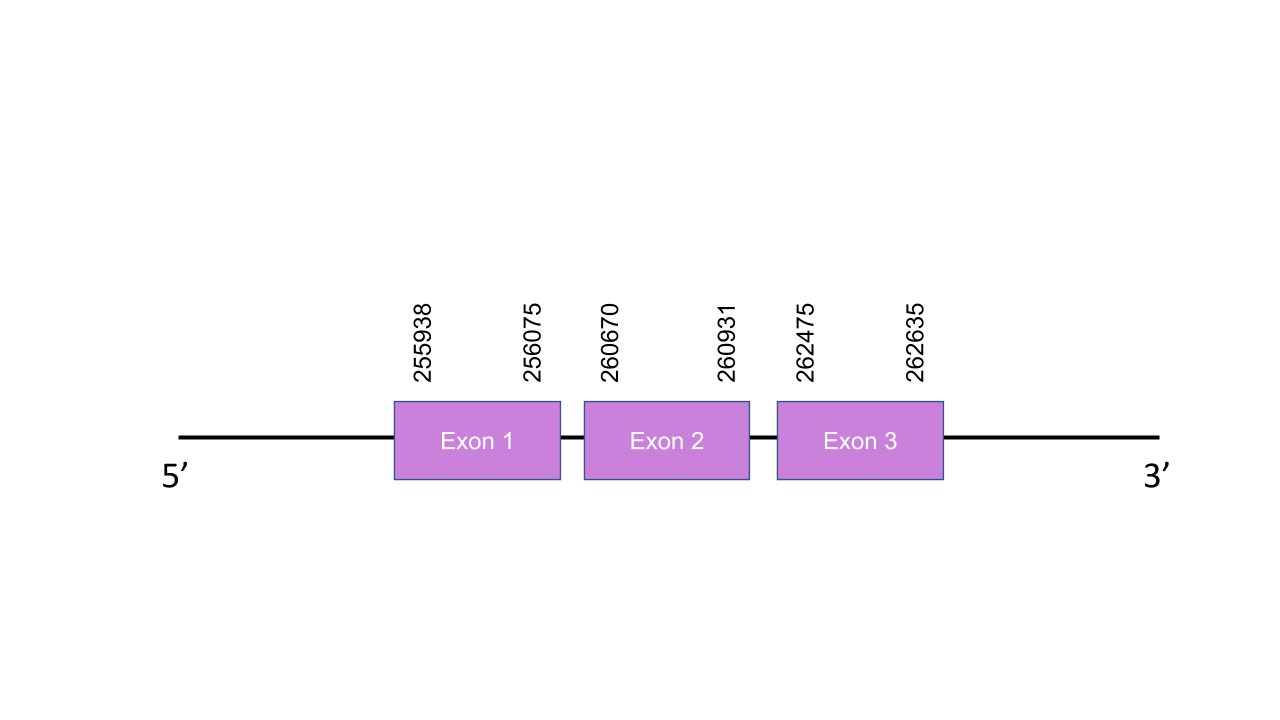
The gene that encodes the protein GPx7 is located in the scaffold PISW01000873.1 between the positions 255995-262635 in the forward strand. Among the 8 scaffolds obtained on the Tblastn, we selected that scaffold since it was the one with the best and highest hit e-value and its 3 alignment fragments has an average identity of 91,00. This gene presents 3 exons predicted with exonerate, located at 255938-256075, 260670-260931 and 262475-262635 in the positive strand. T-coffee shown an excellent aligment with a 995 score between the two species although our predicted protein doesn’t starts with a Methionine but with a Valine(V). Furthermore, it is shown that is a cysteine homolog since it did not have any Sec residue, same as the human protein. Thus, it is a Cys-containing homolog in the two species, since the Sec has been reversed to Cys residue, but the protein might preserve a similar function. Seblastian was unable to found any SECIS elements neither any known selenoprotein.
GPx8
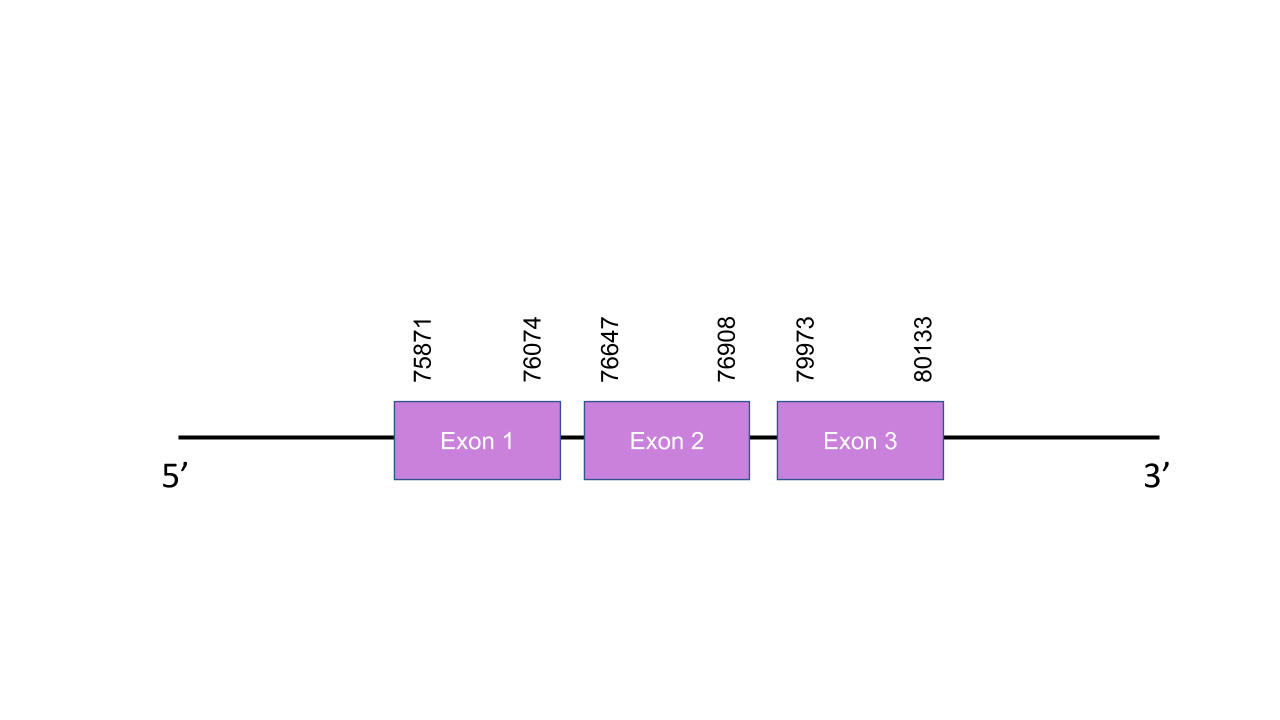
The gene that encodes the protein GPx8 is located in the scaffold PISW01006942.1 between the positions 75871-80133 in the forward strand. Among the 6 scaffolds obtained on the Tblastn, we selected that scaffold since it was the one with the best and highest hit e-value and its 3 alignment fragments has an average identity of 86,734. This gene presents 3 exons predicted with exonerate, located at 75871-76074, 76647-76908, 79973-80133 in the positive strand. T-coffee shown an score of 1000 so it was a perfect aligment between the two species.The predicted protein starts with a Methionine as the human does. Furthermore, it is shown that this is a cysteine homolog since it did not have any Sec residue on either of the two species, but it has many conserved cysteines. Seblastian was able to found a grade B SECIS element between 25550-25635 positions on the negative strand, however this structure had to be dismissed since it was on the opposite strand of our gene.
Iodothyronine deiodinase (DIO)
DIO1
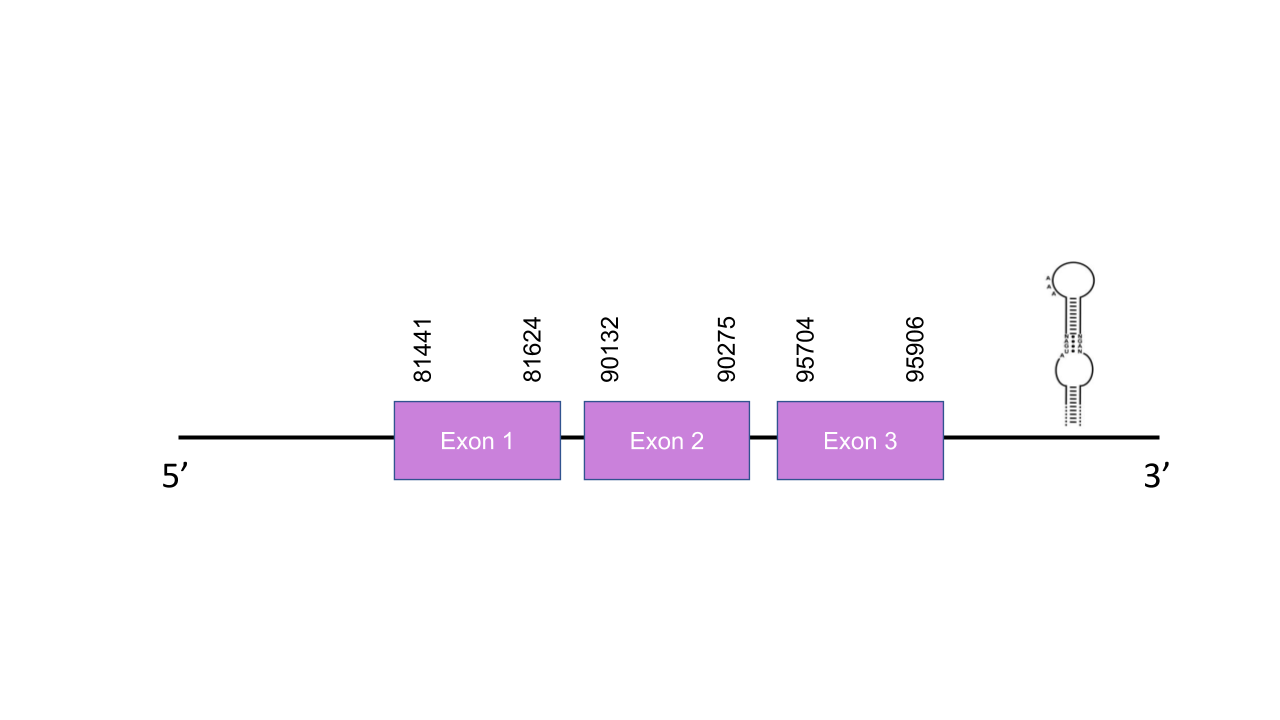
The gene that encodes the protein DIO1 is located in the scaffold PISW01001659.1 in the forward strand (e-value=4.13e-32), between the positions 81441 and 95906. This scaffold was chosen because, although there is another scaffold (PISW01007923.1) with a slightly higher e-value (3.10e-37), the chosen hit showed a higher average identity (83,024). Moreover, a hit in the scaffold PISW01007923.1 was also obtained for the protein DIO3 with a dramatically higher e-value (9.46e-167). This gene has three exons: the first one between the positions 81441 and 81624, the second one between the positions 90132 and 90275, and the third one between the positions 95704 and 95906. There is one SECIS between the positions 52794 and 52864. The T-coffee has shown a result of 999. The selected protein, unlike the human one, does not start with a methionine, it starts with a lysine. The only selenocysteine found in the human protein is conserved in Mungos mungo. Seblastian predicted one known selenoprotein and one grade A SECIS in the forward strand between the positions 98497 and 98567 in the 3’UTR.
DIO2
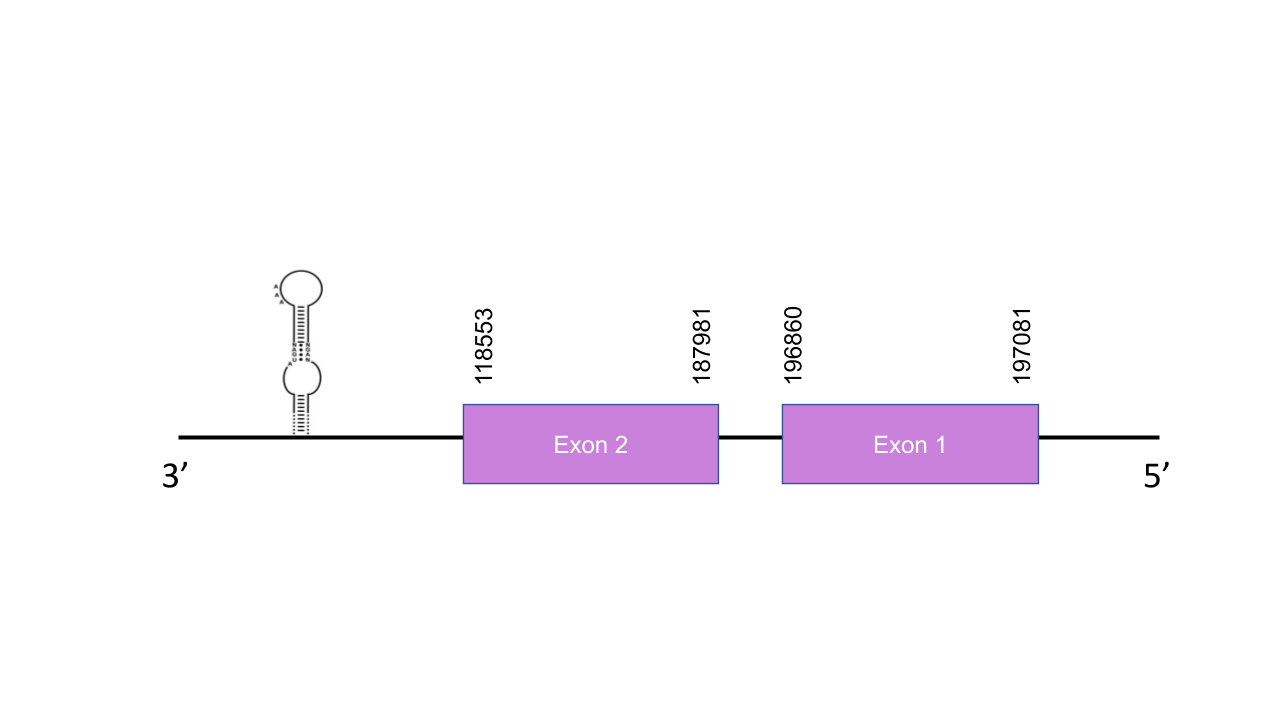
The gene that encodes the protein DIO2 is located in the scaffold PISW01000406.1 between the positions 187981 and 197081 in the reverse strand. Although 4 different hits were obtained, the chosen one had a dramatically higher e-value (1.07e-113) This gene has two exons: exon 1 between the positions 196860 and 197081, and exon 2 between the positions 187981 and 188553. The T-coffee has shown a score of 999. The selected protein starts with a methionine and has only one conserved selenocysteine of the two that are present in the human protein. Seblastian predicted no known protein. However, a grade A SECIS was found between the positions There is one SECIS between the positions 183054 and 183126 in the 5’UTR.
DIO3
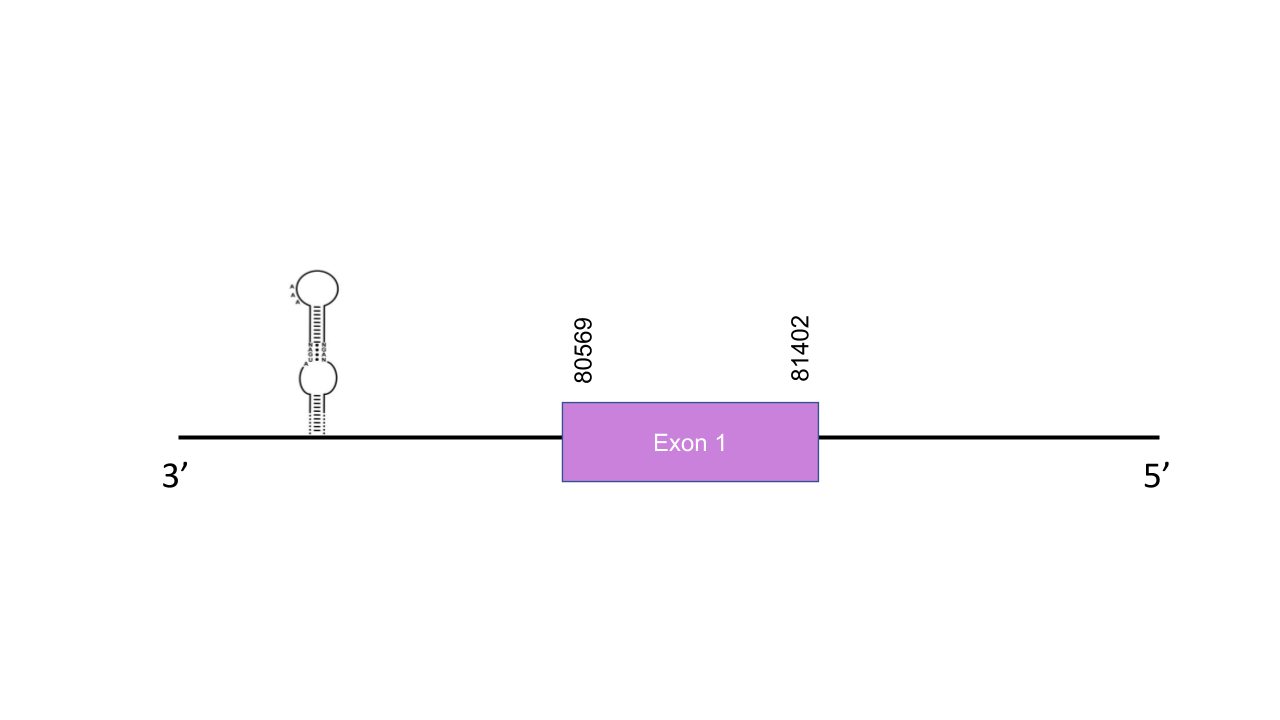
The gene that encodes the protein DIO3 is located in the scaffold PISW01007923.1 between the positions 80569-81366 in the reverse strand. Although 4 different hits were obtained, the chosen one had a dramatically higher e-value (9.46e-167). This gene has one exon between the positions 80569 and 81402. The T-coffee has shown a score of 1000. The selected protein starts with a methionine and the only selenocysteine found in the human protein is conserved. Seblastian predicted one known selenoprotein and one grade A SECIS in the reverse strand between the positions 79897 and 79966 in the 5’UTR. In order to confirm that the proteins have been predicted correctly, a phylogenetic tree was built. As the following figure shows, each query protein (human) is closest it correspondent predicted protein, thus, we can conclude that the prediction is accurate.
Thioredoxin reductases (TXNRDs)
TXNRD1
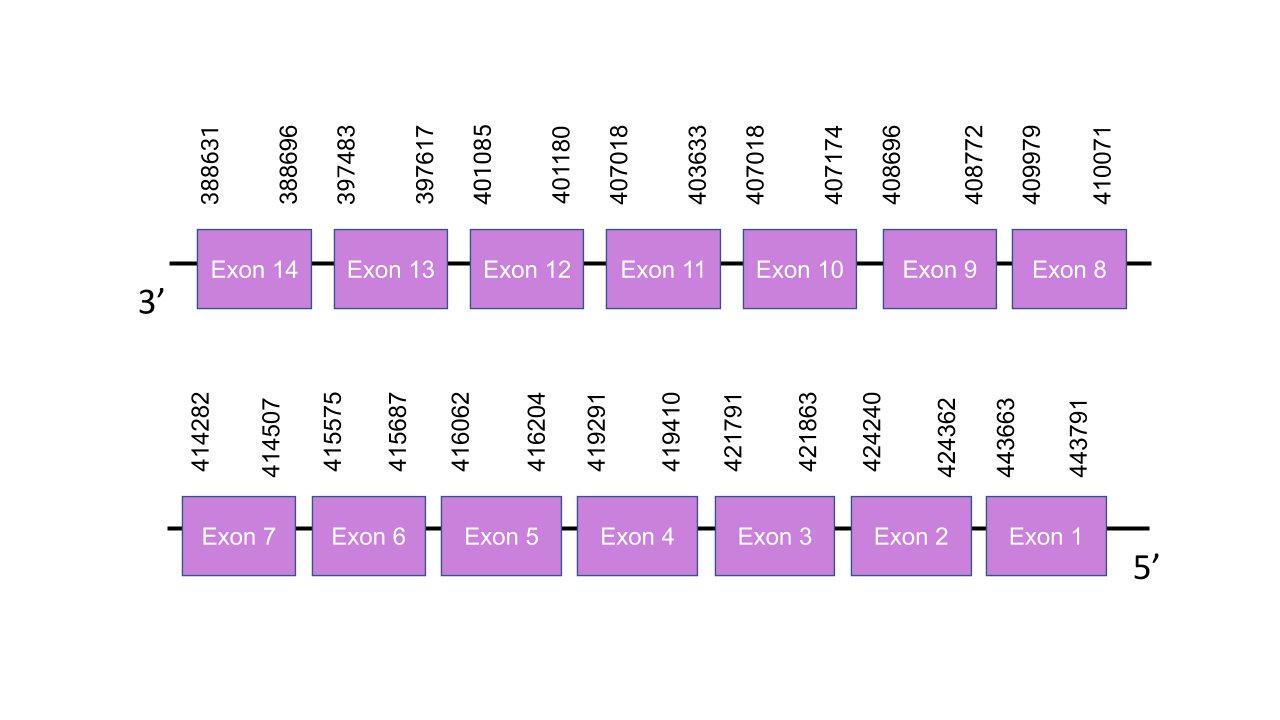
The gene encoding the TXNRD1 protein is located in the scaffold PISW01000907.1, between the positions 414512 and 414270 in the reverse strand. This hit was chosen because it has the highest e-value (3.00e-39). Exonerate enabled to predict fourteen exons: exon 1 is between the positions 443663 and 443791, exon 2 is between the positions 424240 and 424362, exon 3 is between the positions 421791 and 421863, exon 4 is between the positions 419291 and 419410, exon 5 is between the positions 416062 and 416204, exon 6 is between the positions 415575 and 415687, exon 7 is between the positions 414282 and 415687, exon 8 is between the positions 409979 and 410071, exon 9 is between the positions 408696 and 408772, exon 10 is between the positions 407018 and 407174, exon 11 is between the positions 403796 and 403633, exon 12 is between the positions 401085 and 401180, exon 13 is between the positions 397483 and 397617, and exon 14 is between the positions 388631 and 388696. The score obtained with T-coffee is 997. The protein starts with a leucine instead of a methionine. The only selenocysteine found in the human protein is conserved in the predicted protein of Mungos mungo. Seblastian predicted the corresponding protein. One SECIS was found in the 3’UTR of the reverse strand between the positions 388410 and 388325.
TXNRD2
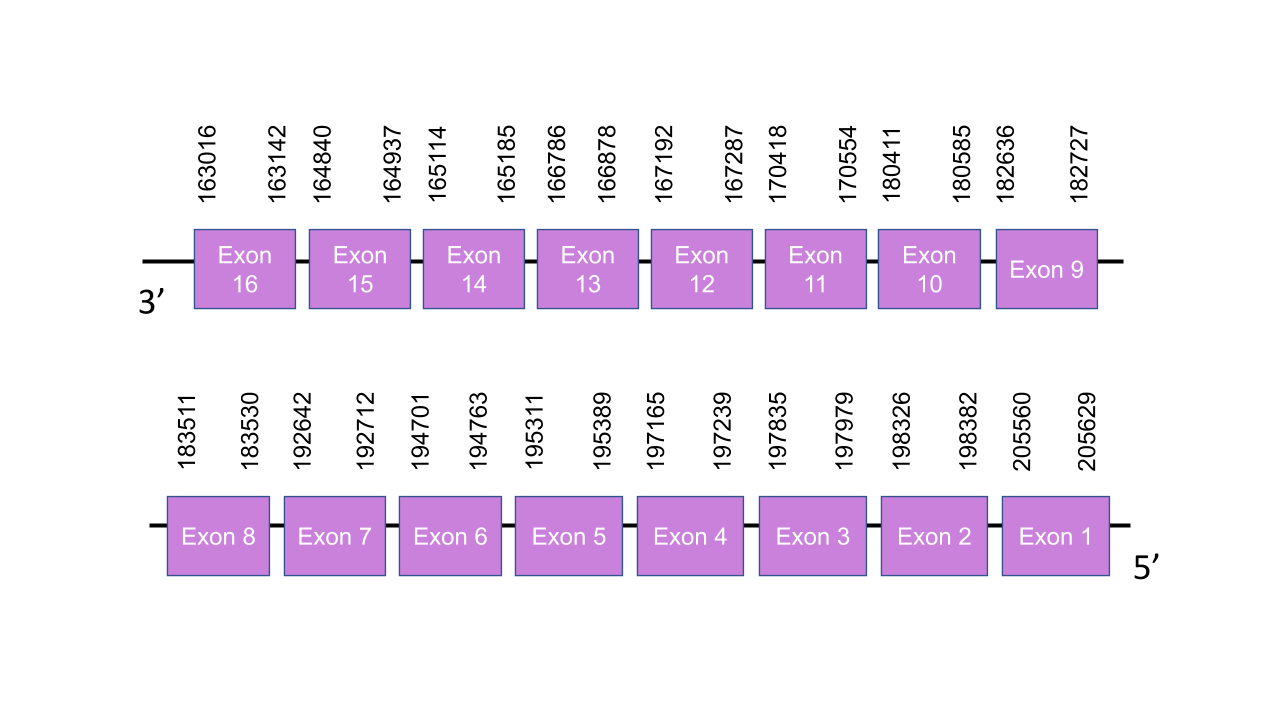
The gene encoding the protein TXNRD2 in Mungos mungo is located in the scaffold PISW01000402.1 between the positions 197992 and 197834 in the reverse strand (e-value = 3.21e-20). This scaffold was chosen because, even though the scaffolds PISW01000027.1 and PISW01000907.1 have a higher e-values (4.88e-28 and 1.57e-21, respectively), the predicted protein in the scaffold PISW01000402.1 shows a higher score in T-coffee than the ones predicted in the other scaffolds. Moreover, the scaffold PISW01000907.1 was assigned to the protein TXNRD1 and the scaffold , which shows a high score in T-coffee. This scaffold assignation can later be confirmed with the phylogenetic tree. Exonerate predicted sixteen exons: exon 1 is between the positions 205560 and 205629, exon 2 is between the positions 198326 and 198382, exon 3 is between the positions 197835 and 197979, exon 4 is between the positions 197165 and 197239, exon 5 is between the positions 195311 and 195389, exon 6 is between the positions 194701 and 194763, exon 7 is between the positions 192642 and 192712, exon 8 is between the positions 183511 and 183530, exon 9 is between the positions 182636 and 182727, exon 10 is between the positions 180411 and 180585, exon 11 is between the positions 170418 and 170554, exon 12 is between the positions 167192 and 167287, exon 13 is between the positions 166786 and 166878 ,exon 14 is between the positions 165114 and 165185, exon 15 is between the positions 164840 and 164937, and exon 16 is between the positions 163016 and 163142. The score obtained with T-coffee is 999. The protein starts with a threonine instead of a methionine. The only selenocysteine found in the human protein is conserved in the predicted protein of Mungos mungo. Seblastian predicted one protein and one grade A SECIS between the positions 161606 and 161680 in the reverse strand.
TXNRD3
The gene encoding the protein TXNRD3 in Mungos mungo is located in the scaffold PISW01007477.1 between the positions 33811 and 33599 in the reverse strand (e-value = 2.25e-31). This scaffold was chosen because, even though the scaffold PISW01000907.1 has a higher e-value (1.51e-32), it shows a higher score in T-coffee than the scaffold PISW01000907.1. Moreover, the scaffold PISW01000907.1 was assigned to the protein TXNRD 1, which shows a high score in T-coffee. This assignation can be confirmed to be correct by the prediction of the Seblastian, which was the corresponding human protein. Exonerate predicted sixteen exons: exon 1 is between the positions 63418 and 63642, exon 2 is between the positions 48264 and 48324, exon 3 is between the positions 46875 and 46984, exon 4 is between the positions 45067 and 45171, exon 5 is between the positions 44771 and 44843, exon 6 is between the positions 42532 and 42651, exon 7 is between the positions 37547 and 37689, exon 8 is between the positions 34597 and 34712, exon 9 is between the positions 33605 and 33830, exon 10 is between the positions 32342 and 32434, exon 11 is between the positions 25925 and 26001, exon 12 is between the positions 25189 and 25345, exon 13 is between the positions 21114 and 21221,exon 14 is between the positions 9430 and 9525, exon 15 is between the positions 6980 and 7114, and exon 16 is between the positions 6701 and 6766. The score obtained with T-coffee is 998. The protein starts with a serine instead of a methionine. The only selenocysteine found in the human protein is conserved in the predicted protein of Mungos mungo. Seblastian predicted the corresponding protein. One SECIS was found in the 3’UTR of the reverse strand between the positions 6513 - 6439.
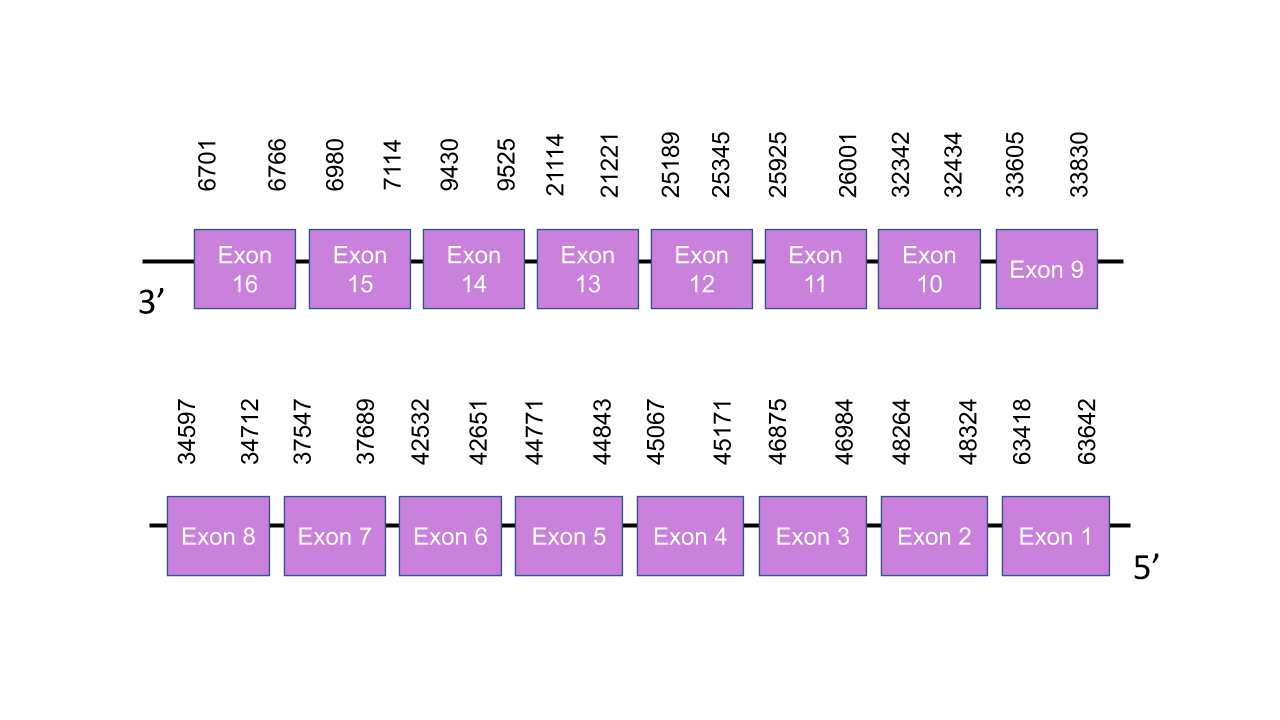
In order to confirm that the proteins have been predicted correctly, a phylogenetic tree was built. As the following figure shows, each query protein (human) is closest it correspondent predicted protein, thus, we can conclude that the prediction is accurate.
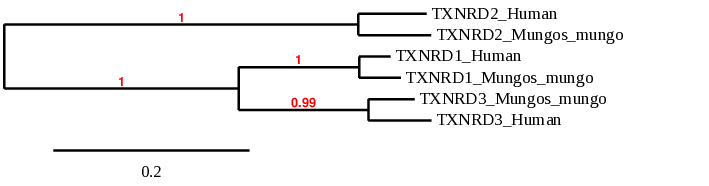
Selenoprotein R or Methionine-R-Sulfoxide Reductase 1 (MSRB)
MSRB1
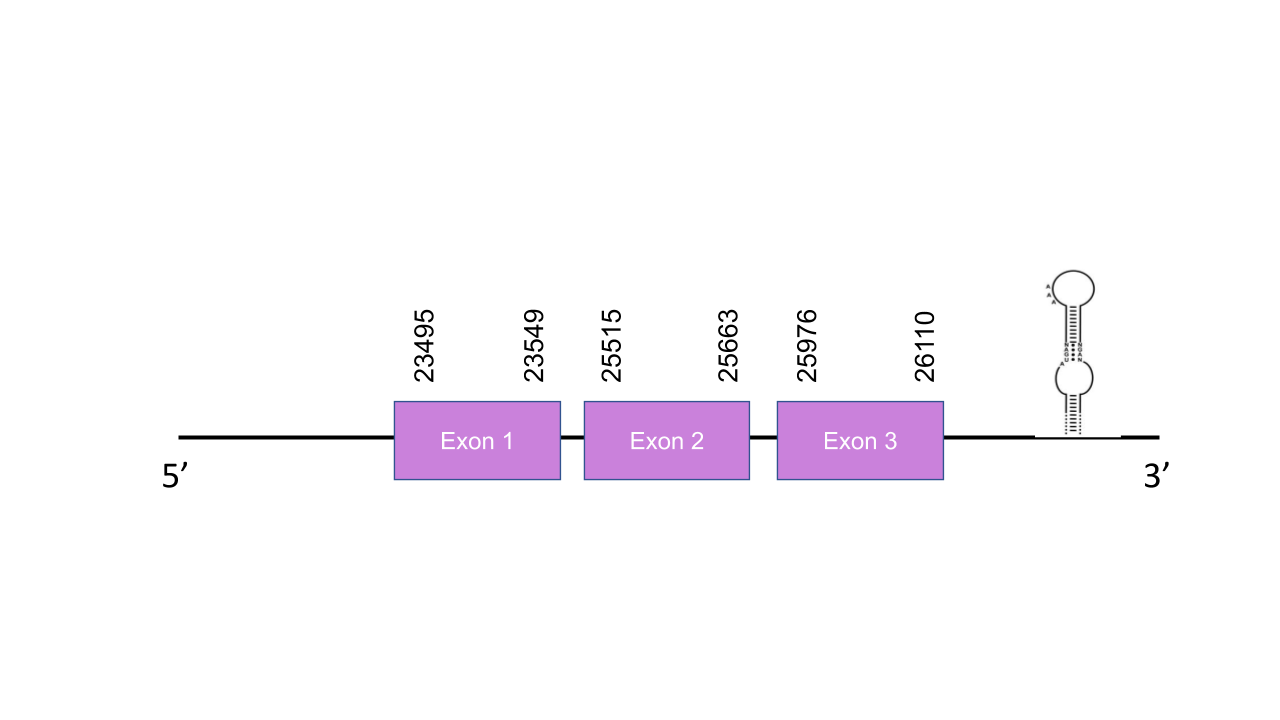
The gene that encode for this protein is located in the scaffold PISW01009745.1. Its is in the forward strand between positions 23495 and 26110 and has got an e-value of 2.02e-23. The gene has got three exons located in the next positions: 23495-23549, 25515-25663, 25976-26110. The T-coffee shows a score of 1000 which is a perfect alignment. As expected, MSRB1 is a selenoprotein presenting a Sec residue in the same position as in the human sequence. Seblastian predicted one SECIS element located in the forward strand between positions 28298 and 28368 at the 3’ end of the strand. It is grade A.
MSRB2
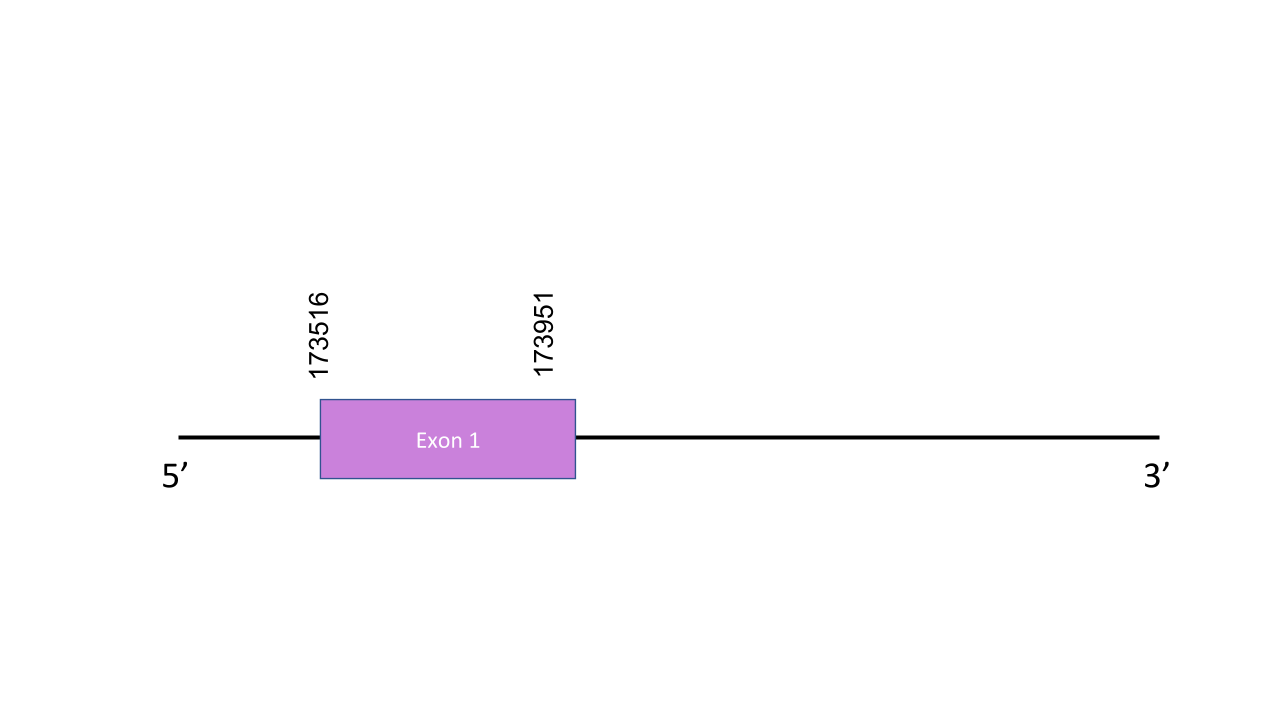
MSRB2 The gene that encode this protein is located in the scaffold PISW01000008.1 which is in the forward strand and positions 173604-173939. It has got only one exon located between 173516 and 173951. About the T-coffee, it has a score of 792. In this case we are talking about a Cys-homologue. There is no coincidence between Sec residues in human and Mungos mungo sequences. Seblastian could not predict any SECIS element nor protein.
MSRB3
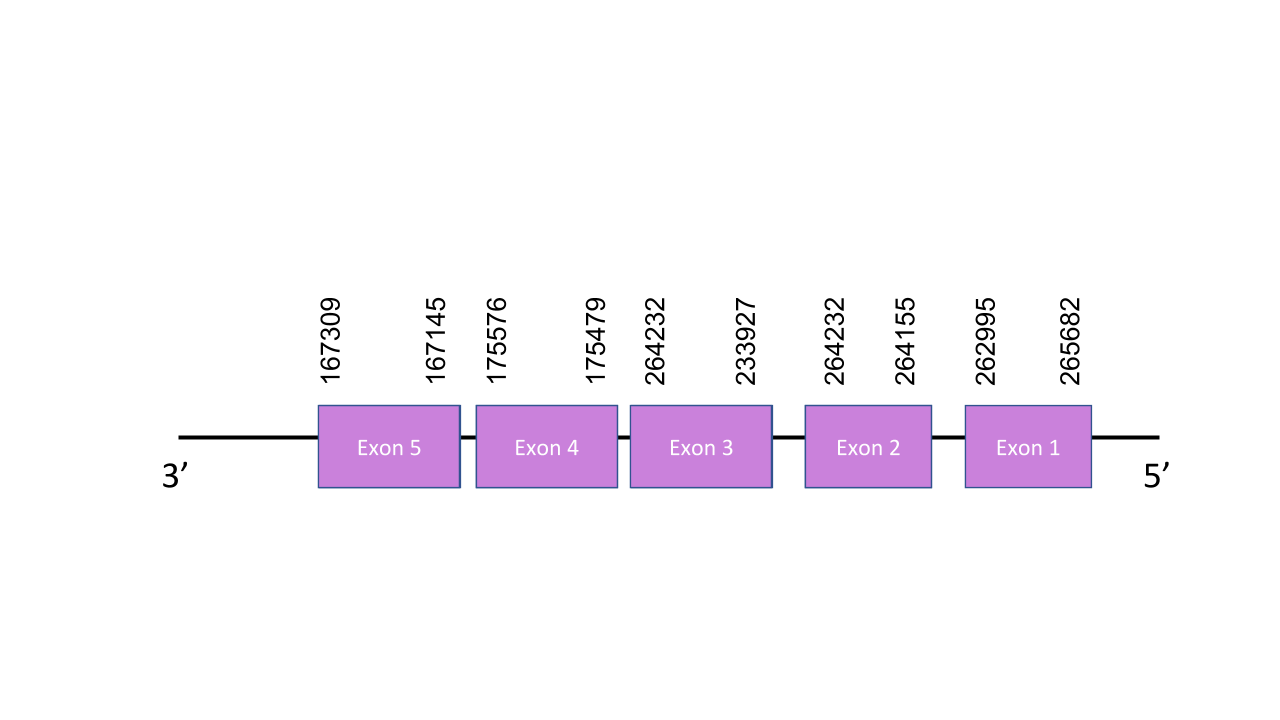
The gene that encode MSRB3 protein is located in the scaffold PISW01002117.1 between positions 167145 and 265962. It has got five exons which are located in positions: 265682-262995, 264155-264232, 233927-264232, 175479-175576, 167145-167309. T- coffee shows a score of 991 which is a value that indIcates almost a perfect alignment. There are no Sec residues but Cys. MSRB3 is a Cys-homologue. Seblastian predicted two SECIS elements, both of grade B. However just one of them is in the reverse strand so the other one cannot be valid. About the one in the reverse strand, it is grade B and located between positions 195302 and 195214. Taking into account the position of the one in the reverse strand we can say there are no SECIS elements.
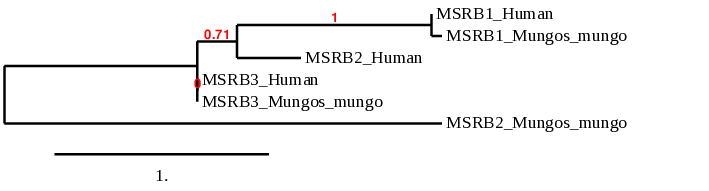
The phylogeny tree of MSRB family correlates with the results obtained. We can see that MSRB1 is highly conserved in Mungo mungo. From another hand MSRB2 is the one which shows more distance between the two especies. In the case of MSRB3 which is the Cys-homologue we can see that it is more conserved.
Methionine sulfoxide reductases A (MsrA)
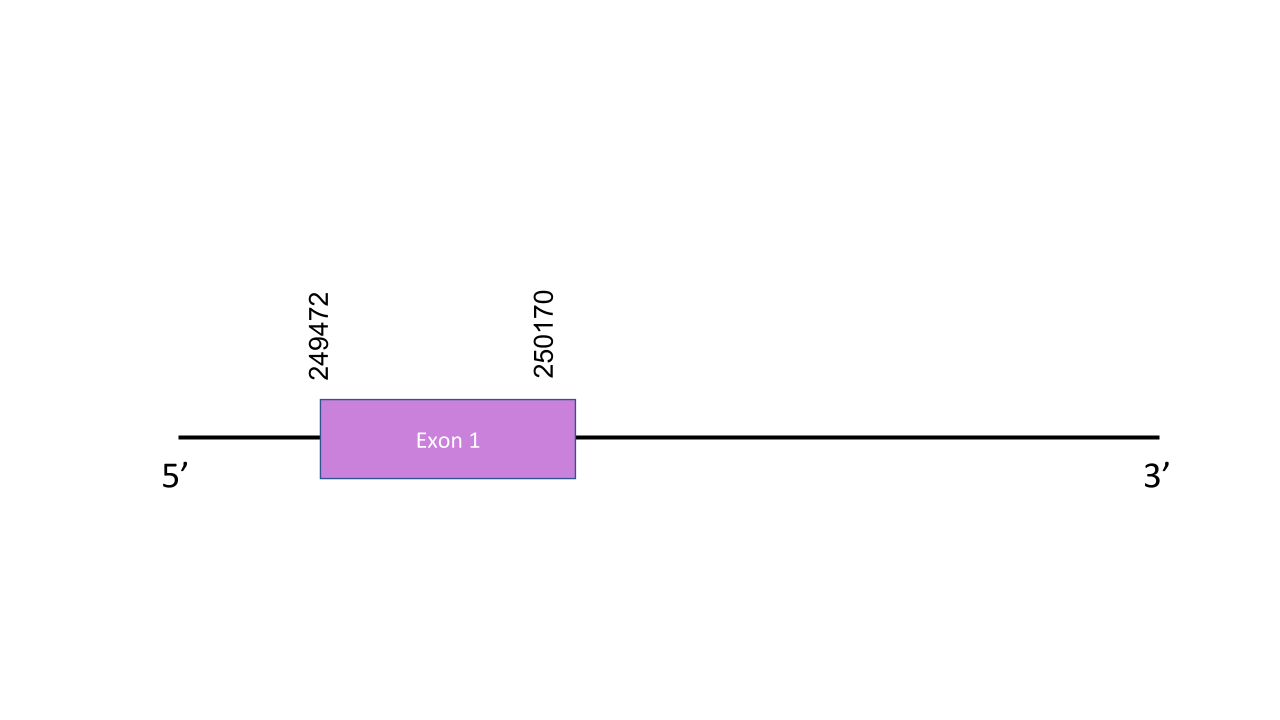
The gene that encodes the protein MsrA is located in the scaffold PISW01000451.1 between the positions 249472 and 250170 in the forward strand. This scaffold was the one with the highest e-value: 3.97e-134. This gene has just one exon and it is located between positions 173516 and 173951. The T-coffee shows a score of 998 which almost perfect. Nevertheless observing the T-coffee alignment we can see there is no Sec residue but Cys. It results that MsrA is a Cys-homologue. Seblastian did not predict any SECIS nor protein.
15-kDa selenoprotein (Sel15)
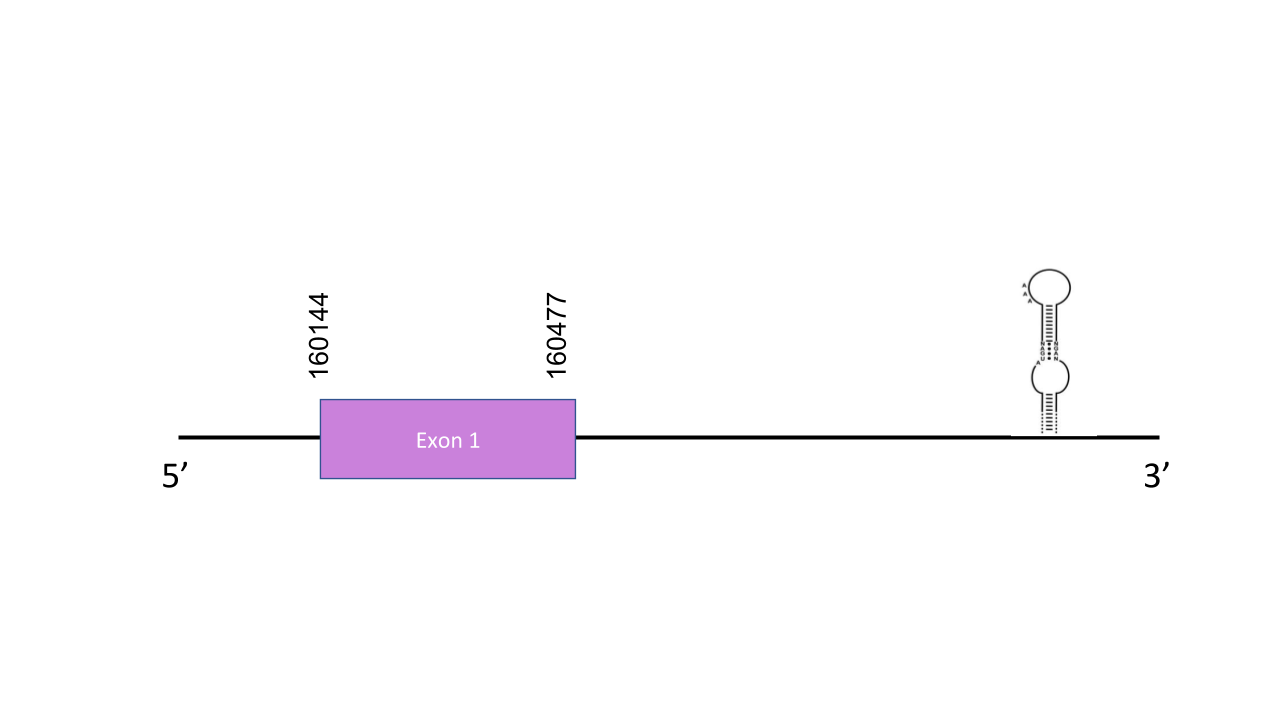
The gene that encodes the protein Sel15 is located in the scaffold PISW01002128.1 between the positions 160022 and 160477 in the forward strand. Among the other scaffolds this was chosen because it has the highest e-value: 5.85e-26.This gene has just one exon and it is located between positions 160144 and 160477. T-coffee shows a very good score since it has a value of 927. It contains a Sec residue so, as expected Sel15 is a selenoprotein. It has two SECIS elements, which were predicted by Seblastian, but just was one is in the positive strand so correlates with the scaffold which is in the forward strand. The SECIS (grade A) is located between positions 161058 and 161135, at the 3’ end of the forward strand.
Selenoprotein I
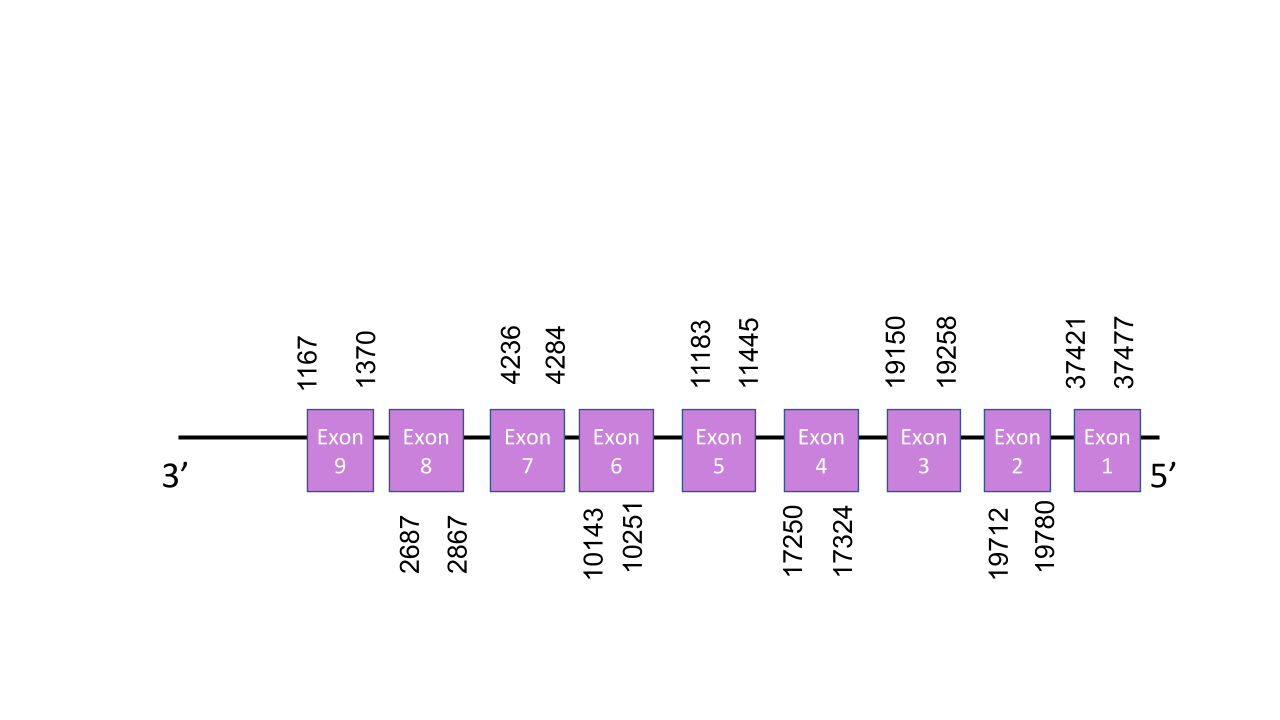
The gene that encodes the protein SelI is located in the scaffold PISW01003253.1 between the positions 1134 and 19783 in the reverse strand. This was the scaffold selected because it was the one with the best e-value: 2.52e-34. The gene has got 9 different exons which are located in the reverse strand and have the next positions:37421-37477, 19712-19780, 19150-19258, 17250-17324, 11183-11445, 10143-10251, 4236-4284, 2687-2867, 1167-1370. T-coffee shows a score of 996 which seems very good since it is near 1000. The alignment does not show any Sec residue for Mungos mungo but a gap. This might mean that Mungos mungo’s genome was not correctly annotated. There was found just on SECIS element of grade B located between positions 39849 - 39920 but it is located in the forward strand so it cannot be valid.
Selenoprotein M
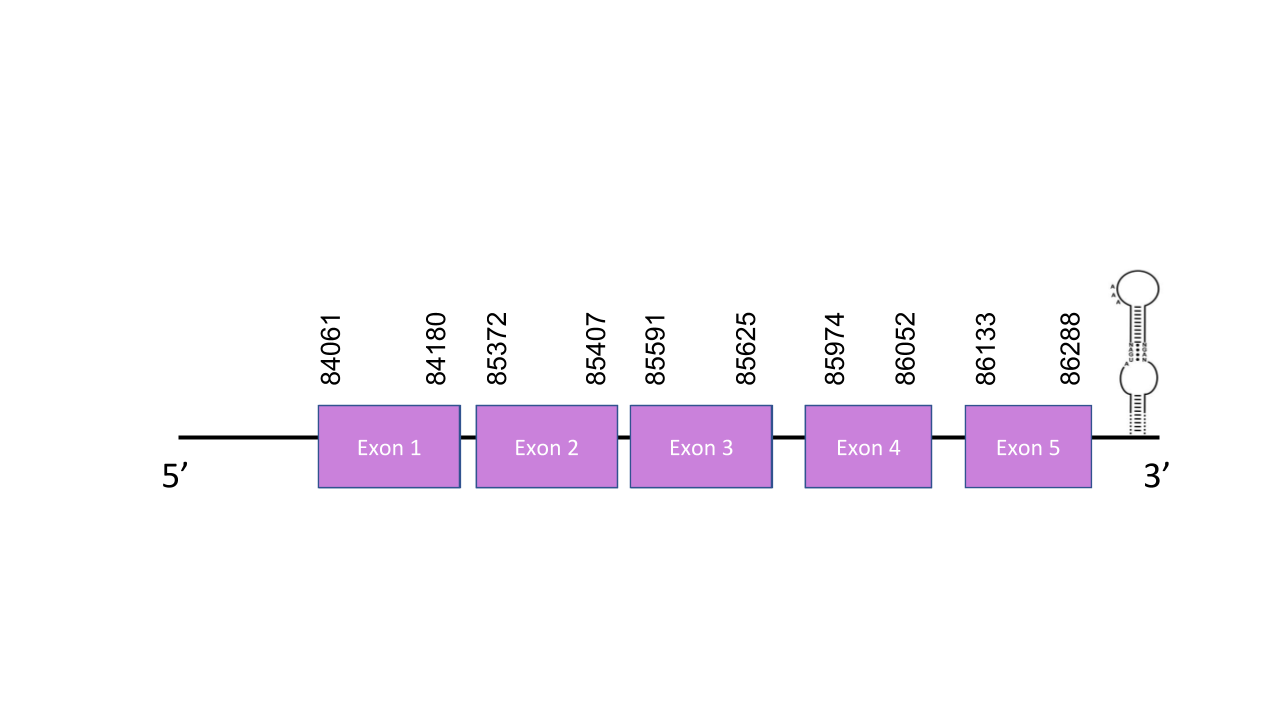
The gene that encodes this protein is located in the scaffold PISW01004272.1 between the positions 85975 and 86288 in the forward strand. This scaffold, which was the only one obtained has got an e-value of 2.68e-25. SelM gene has got five exons located in positions: 84061-84180, 85372-85407, 85591-85625, 85974-86052, 86133-86288. T-coffee shows a score of 996 which means there has been a good alignment. It is shown a Sec residue confirming that SelM is a selenoprotein. In this case we might be talking about a duplication since there were two same hits in the same scaffold with the same e-value. Seblastian predicted two SECIS elements and none proteins. Both of the SECIS were found in the forward strand as well as the scaffold. However the first SECIS was grade B and was located in positions 94795 - 94876 and the second SECIS was grade A and located between positions 50202 and 50275. Taking into account these positions we can only take like valid the one which es grade B.
Selenoprotein N
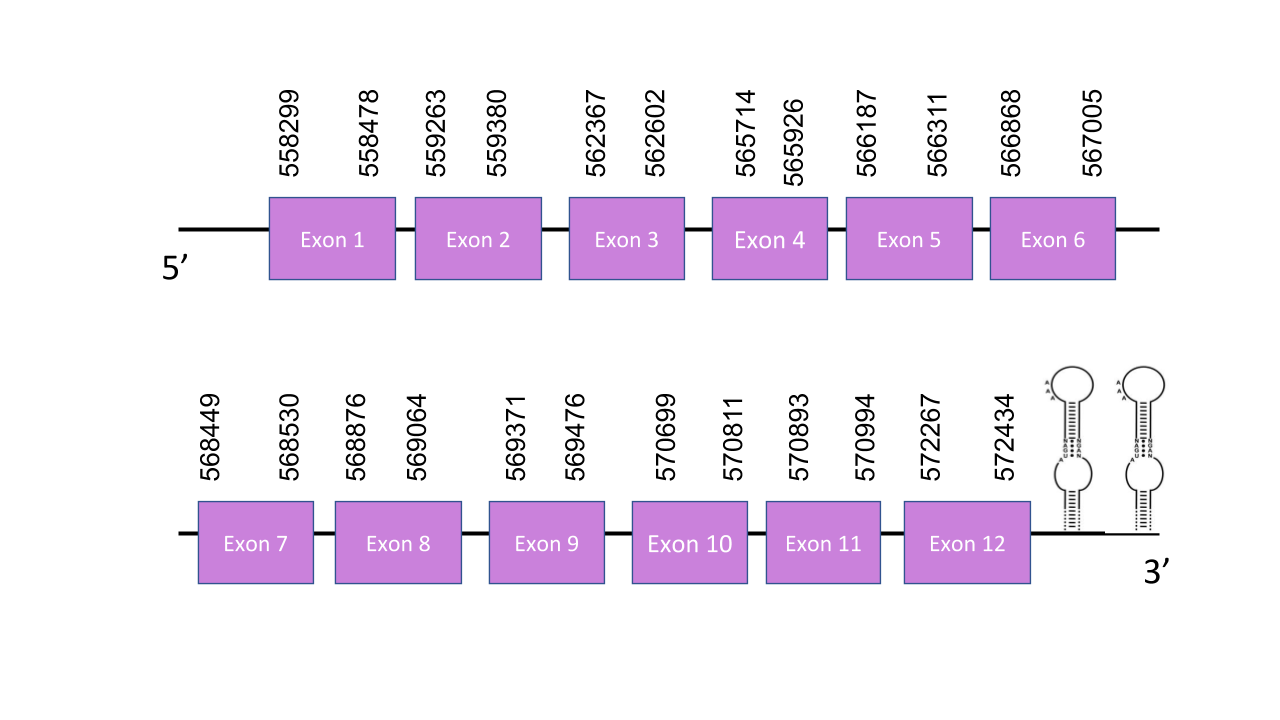
The gene that encodes this protein is located in the scaffold PISW01000482.1 in the forward strand. This was the one chosen because it has got the best e-value: 2.95e-29. The gen has got 12 exons which are located in the next positions: 558299- 558478, 559263-559380, 562367-562602, 565714-565926, 566187- 566311, 566868- 567005, 568449-568530, 568876-569064, 569371-569476, 570699-570811, 570893-570994, 572267-572434. The score obtained by T-coffee has a value of 987 which is a high one meaning that there was a good alignment. The alignment shows a Sec residue in the same position as the human sequence confirming SelN is a selenoprotein. Seblastian predicted three SECIS elements, all of them in the forward strand. The first one was grade A and was located between positions 573507 and 573574. The second SECIS element was grade B and located between positions 570736 and 570820. Lastly, the third one was grade B and located between 573108 and 573181. Therefore there are just two of them which are valid and located in the 3’ end of the forward strand.
Selenoprotein O
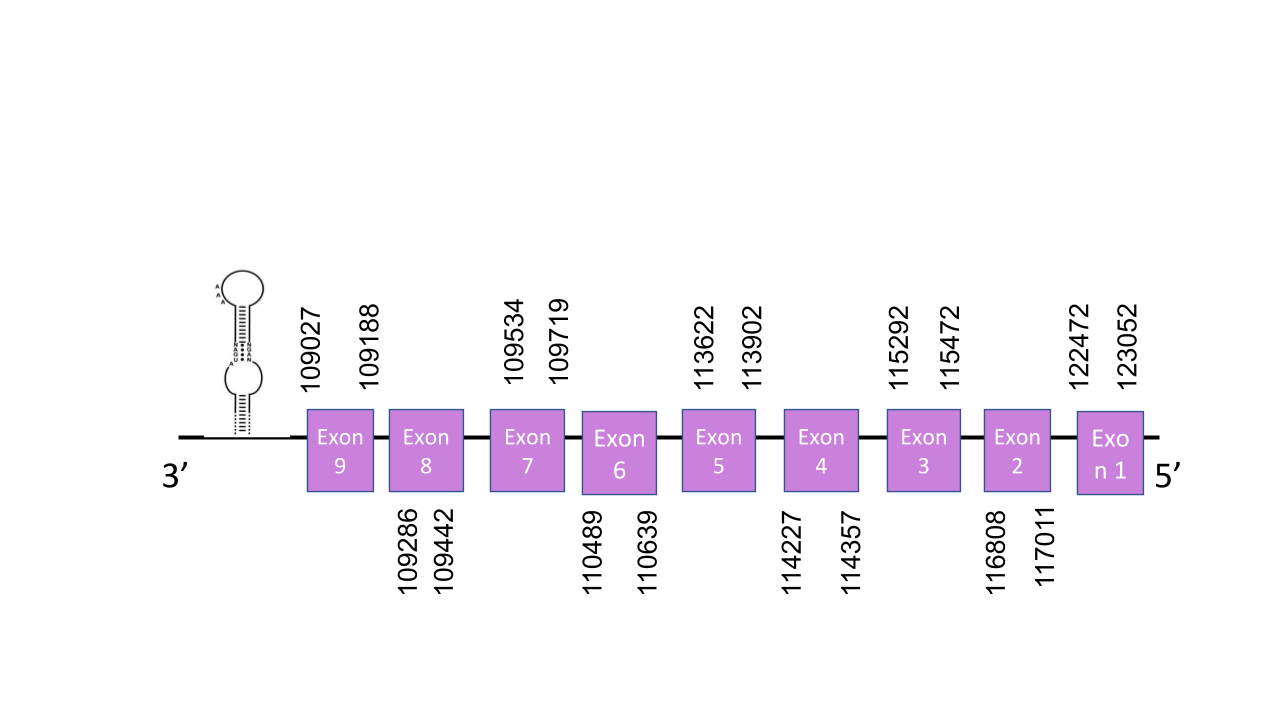
The gene that encode for SelO protein is located in the scaffold PISW01003802.1 which is in the reverse strand. This was the scaffold with the highest e-value: 4.92e-56 The gene has got 9 exons which are located in positions: 122472-123052, 116808-11701, 115292-115472, 114227-114357, 113622-113902, 110489-110639, 109534-109719, 109286-109442, 109027-109188. T-coffee shows a score of 991 which means that it is almost a perfect alignment. It is shown a Sec residue in the same position as the human sequence meaning that SelO is a selenoprotein. Seblastian predicted two SECIS elements but just one of them was in the reverse strand. This one is grade A and it is located between positions 108935 and 108860 at the 3’ end of the reverse strand. In this case Seblastian did predict the protein.
Selenoproteins P
The gene that encode this protein is located in the scaffold PISW01002351.1 which is in the reverse strand. The scaffold was chosen due to its high e-value: 2.98e-37. This gene has got four exons which are located between positions: 212296-212498, 210738-210950, 209075-209192, 206383-207006. It has a score in T-coffee of 980 which is quite good. The alignment shows various Sec residues that match with the ones shown in the human sequence. As expected SelP is a selenoprotein. besides there are shown extra Sec residues in the Mungos mungo sequence. Seblastian predicted three SECIS elements and three proteins. One of the SECIS was located in the forward strand so it cannot be valid. About the other two, the first one was grade B and was located in positions 205671 - 205606. The other one was grade A and was located between positions 206124 and 206042. Both of them in the 3’ end of the reverse strand.
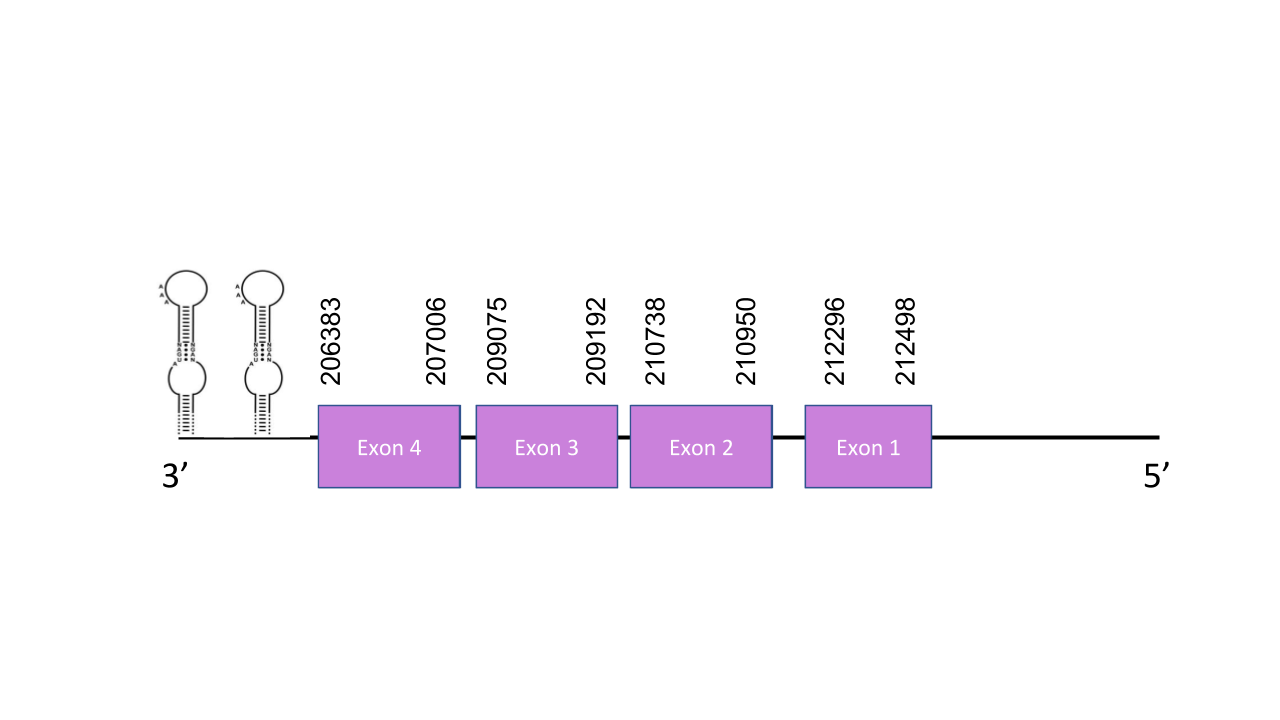
Selenoproteins K and S
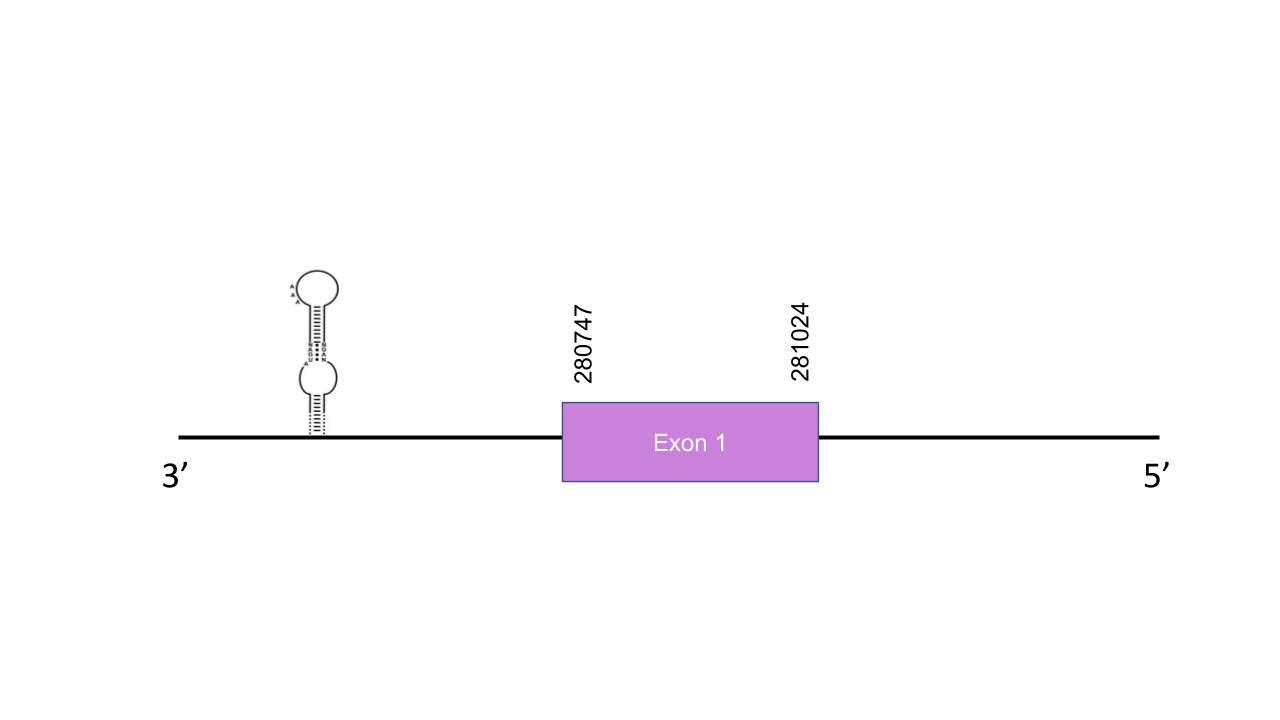
Selenoprotein K
The gene that encodes the protein SelenoK is located in the scaffold PISW01000182.1 between the positions 280789-280953 in the reverse strand. This gene has one exon as predicted with exonerate, located at 281024-280747. We can observe that this gene is duplicated in Mungos mungo, since two hits in the same scaffold and equal e-values were found after running tblastn. Seblastian was not able to predict a known selenoprotein from this Mungos mungo sequence but three grade-B SECIS structures were found between the positions 245635-245674, 330586-330662 and 272044-272128 of the reverse strand. Still, only one of these SECIS (245635-245674) is considered viable, since it is the only one found after the gene. The predicted mongoose selenoprotein has a selenocysteine residue and a high score and e-value when aligned with its human homologue, thus, it is likely that SelK is found in the Mungos mungo genome.
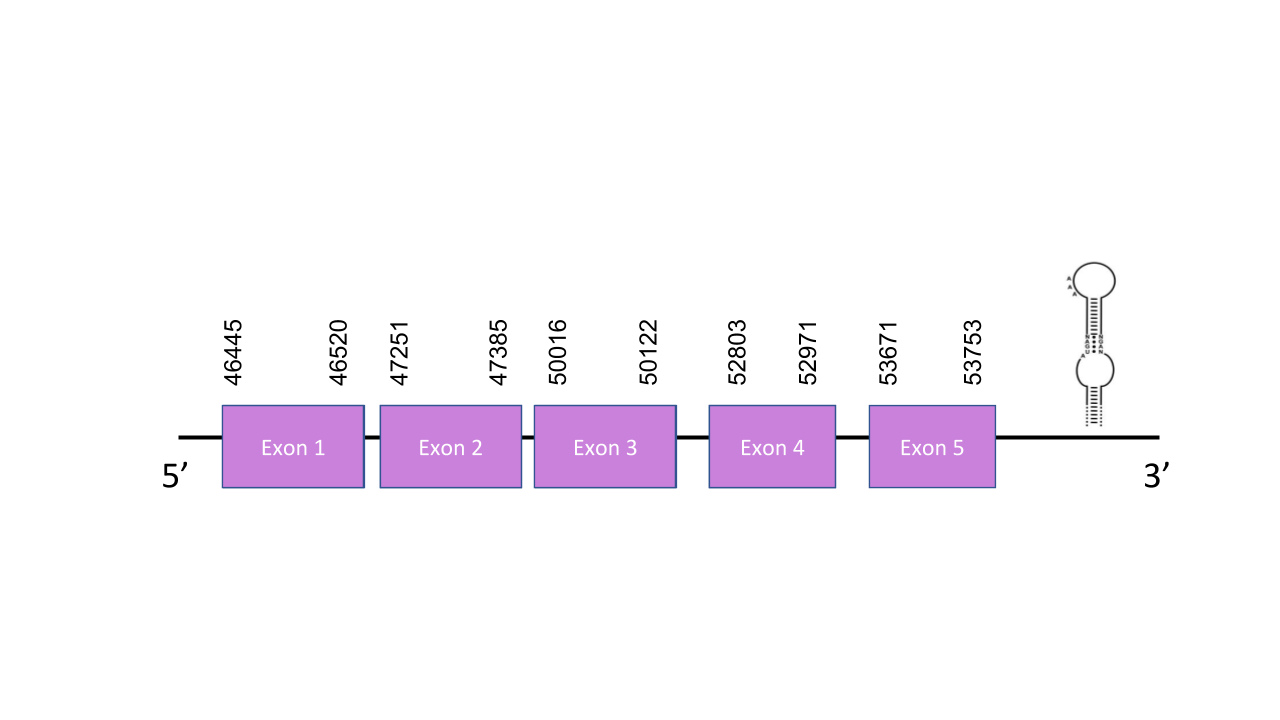
Selenoprotein S
The gene that encodes the protein SelenoS is located in the scaffold PISW01006009.1 between the positions 46445 and 52973 in the forward strand. This gene has five exons as predicted with exonerate, located at 46445-46520, 47251-47385, 50016-50122, 52803-52971, 53671-53753. Seblastian was able to predict a known selenoprotein from the Mungos mungo sequence and a grade A and a grade B SECIS structures were found between the locations 54109-54188 and 9564-9640, respectively, of the forward strand. the grade B SECIS is not a viable structure, since it is located before the SelenoS gene (close to the 5’ end), the grade B one, on the other hand, is viable and located towards the 3’ end of the strand. A 90% identity, a perfect alignment score, a very low e-value and the presence of a selenocysteine residue in the predicted mongoose protein suggest that it is very likely for the mongoose to have the selenoprotein S in its genome.
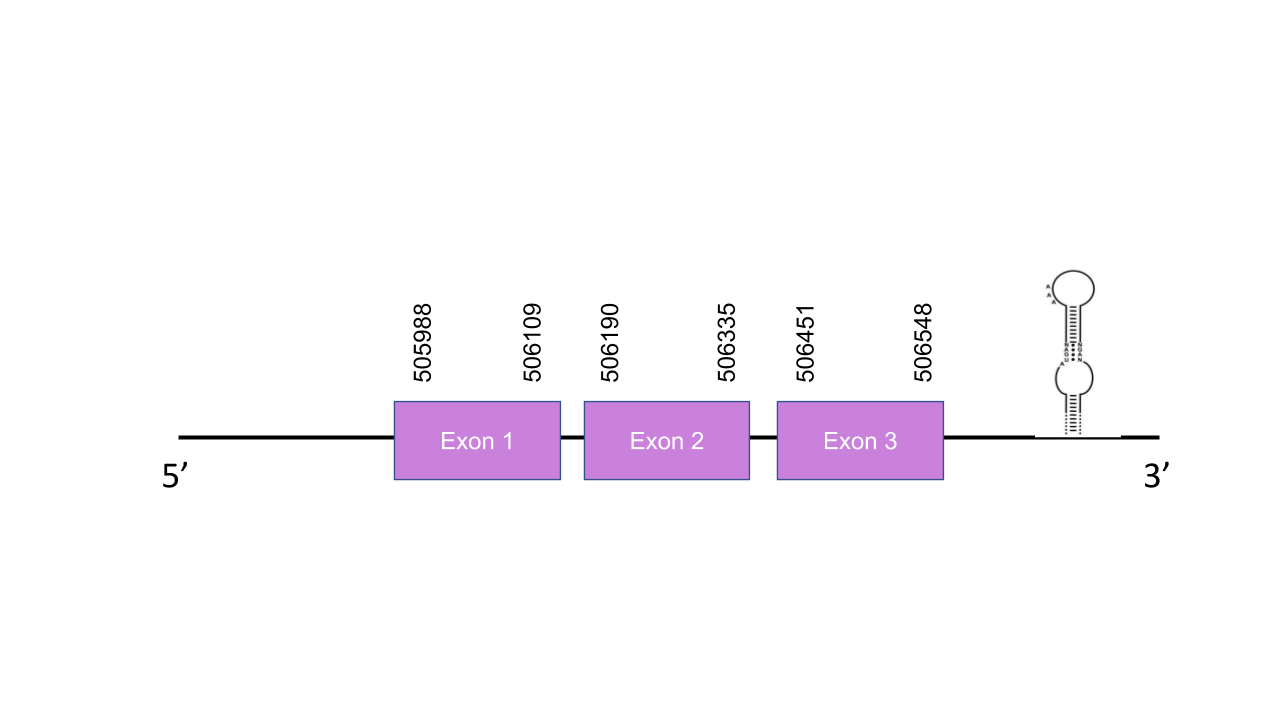
Selenoproteins H, T, V and W
SelH
The gene that encodes the protein SelenoH is located in the scaffold PISW01000182.1 between the positions 505988 and 506548 in the forward strand. This gene has three exons as predicted with exonerate, located at 505988-506109, 506190-506335 and 506451-506548. We can observe that this gene is triplicated in Mungos mungo, since three hits in the same scaffold and equal e-values were found after running tblastn. Seblastian was able to predict a known selenoprotein from the Mungos mungo sequence. Also, a grade A SECIS structure was found between the locations 507129 and 507195 of the forward strand -this is a viable SECIS structure, since it is located in the same strand of our predicted gene and after it. After aligning the predicted protein with the human selenoprotein SelH, we obtained a perfect score of 1.000/1.000. We also were able to find a selenocysteine residue. Altogether, all of this results point towards the fact that SelenoH is a selenoprotein found in the Mungos mungo genome.
SelT
The gene that encodes the protein SelenoT is located in the scaffold PISW01003014.1 between the positions 98676 and 98819 in the forward strand. This hit was chosen because it exhibits the highest e-value (2.63e-21). This gene has four exons: the first one between the positions 49354 and 49427, the second one between the positions 50002 and 50128, the third one between the positions 52118 and 52205, and the fourth one 53730 and 53851. The t-coffee has shown a score of 1000. The predicted protein starts with a methionine. There are no selenocysteines in the human protein nor in the predicted Mungos mungo protein. Seblastian predicted no known protein. However, a forward grade A SECIS was found between the positions 103100 and 103173 in the 3’UTR.
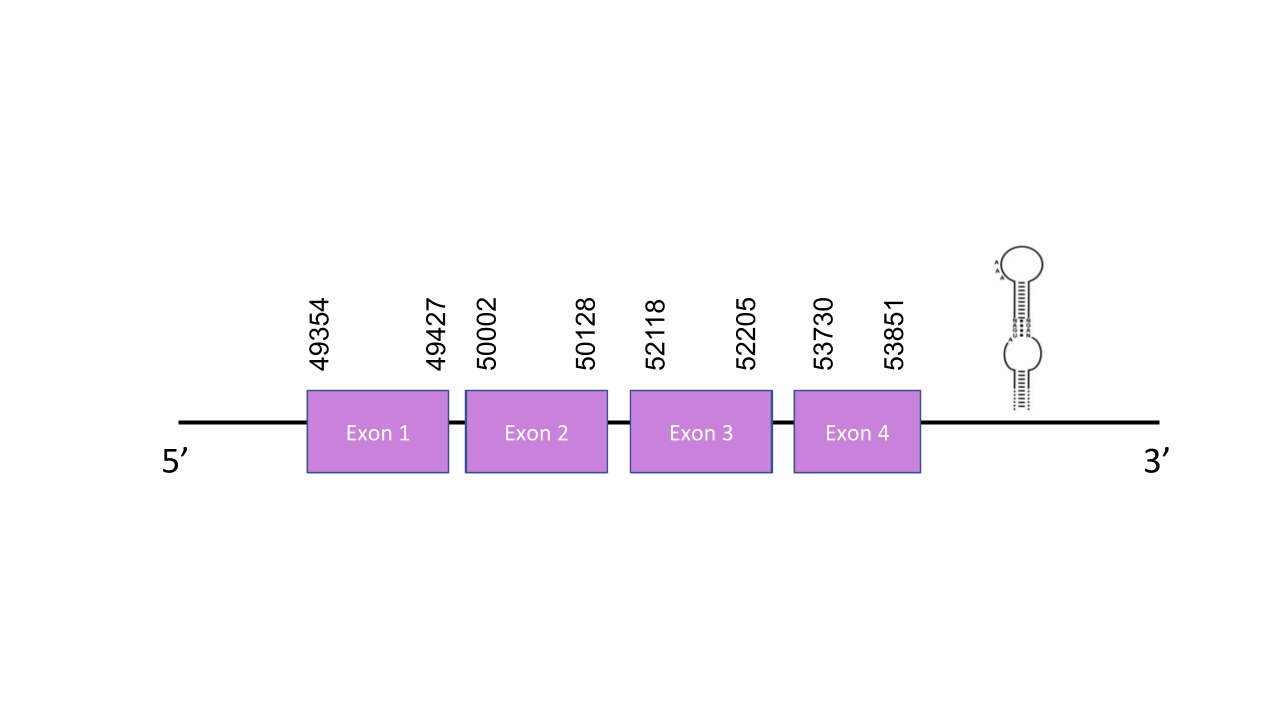
SelW1
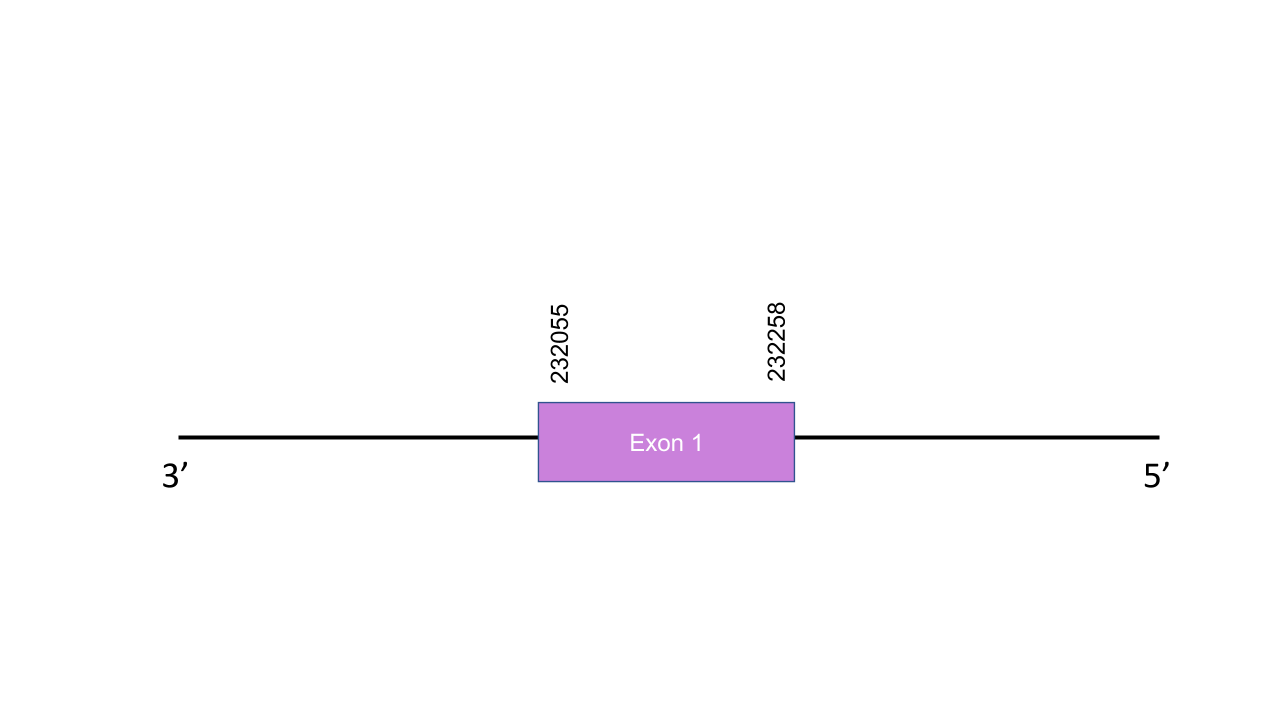
The gene that encodes the protein W1 is located in the scaffold PISW01002260.1 between the positions 232103 and 232234 in the reverse strand. This scaffold was chosen because it had the highes e-value (2.53e-13). This gene has one exon between the positions 232055 and 232258. The T-coffee has shown a score of 987. The selected protein starts with a tyrosine instead of a methionine and the only selenocysteine found in the human protein is not present in the predicted protein, there is a gap. This might be due to a misannotation of the genome of Mungos mungo. For this reason it cannot be confirmed whether the protein W1 is a selenoprotein, a cysteine-containing homologous or other amino acid-containing homologous. Seblastian has not predicted any known protein or SECIS.
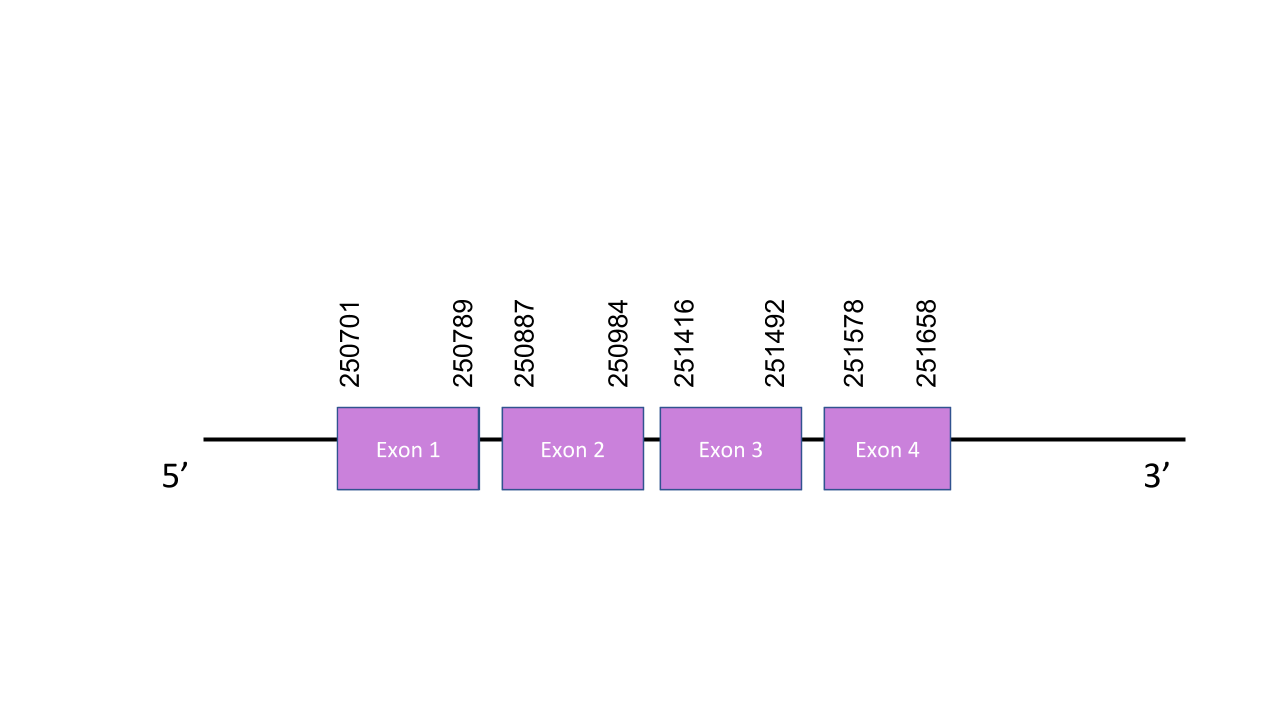
SelW2
The gene that encodes the protein W1 is located in the scaffold PISW01002260.1 in the forward strand. As two hits have been found in the same scaffold with identical e-values (1.31e-17), it can be concluded that this gene is duplicated. This gene has four exons: the first one between the positions 250701 and 250789, the second one between the positions 250887 and 250984, the third one between the positions 251416 and 251492, and the fourth one 251578 and 251658. The t-coffee has shown a score of 1000. The predicted protein starts with a methionine. There are no selenocysteines in the human protein nor in the predicted Mungos mungo protein. No SECIS or selenoproteins have been predicted with Seblastian.
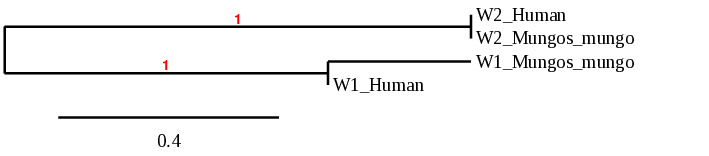
In order to confirm that the proteins have been predicted correctly, a phylogenetic tree was built. As the following figure shows, each query protein (human) is closest it correspondent predicted protein, thus, we can conclude that the prediction is accurate.
SelV
According to the results obtained with tblastn, the gene encoding the Selenoprotein V could be found in the scaffolds PISW01156266.1, PISW01033597.1 and PISW01033137.1. Two hits with the same e-value were obtained for each scaffold (the e-values are 1.10e-09, 2.89e-08 and 3.37e-08, respectively) and those e-values are very similar. As the hits for each scaffold had the same e-value and did not overlap, it can be assumed that the protein is duplicated in each of the scaffolds. The gene has two exons, which in the scaffold PISW01156266.1 are located between the positions 6 and 80 (exon 1) and the positions 190 and 264 (exon 2) and in the scaffolds PISW01033597.1 and PISW01033137.1 are found between the same positions 301 and 363 (exon 1) and 120 and 194 (exon 2). T-coffee shows very high scores for each predicted protein: 1000 for the one in the scaffold PISW01156266.1 and 999 for the ones in the scaffolds PISW01033597.1 and PISW01033137.1. However, the three predictions show a considerable gap previous to the aligned region. The predicted protein in scaffold PISW01156266.1 starts with a glutamic acid instead of methionine and the predicted proteins in the scaffolds PISW01033597.1 and PISW01033137.1. start both with a serine. The only selenocysteine present in the human protein is not conserved in any of the predicted copies of the protein. In the three cases there is a gap in the alignment of the predicted protein with the human selenocysteine. This might be due to a misannotation of the genome of Mungos mungo. For this reason it cannot be confirmed whether the Selenoprotein V is a selenoprotein, a cysteine-containing homologous or other amino acid-containing homologous. there is a gap. This might be due to a misannotation of the genome of Mungos mungo. For this reason it cannot be confirmed whether the protein W1 is a selenoprotein, a cysteine-containing homologous or other amino acid-containing homologous. Seblastian did not predict any protein or SECIS. Given the exposed results, it can be estimated that there are three duplicated copies of the protein. However, this cannot be concluded yet as the results are not consistent enough.

Selenoproteins U
SelU1
The gene that encodes the protein SelenoU1 is located in the scaffold PISW01000505.1 between the positions 113651 and 122251 in the forward strand. This gene has five exons as predicted with exonerate, located at 113651-113828, 114934-115025, 117880-118020, 118705-118869 and 122141-122251. Seblastian was not able to predict a known selenoprotein from this Mungos mungo sequence. Also, a grade B SECIS structure was found between the locations 118164 and 118234 of the forward strand -this is not a viable SECIS structure, since it is located in the same strand of our predicted gene but it is before it. Seblastian is a highly automatized algorism, which makes it less sensitive than manual predictions. After aligning the predicted protein with the human selenoprotein SelU1, we obtained a perfect score of 1.000/1.000. We found out that there was no selenocysteine residue in the mongoose protein, still, this was not surprising since, the human one did not have selenocysteine either. Both of those proteins had cysteine instead, making them cysteine homologues. In spite of that, even though it is likely that Mungos mungo has SelU1, we can not claim for sure that it is found in its genome, due to a lack of SECIS and predicted proteins through Seblastian.
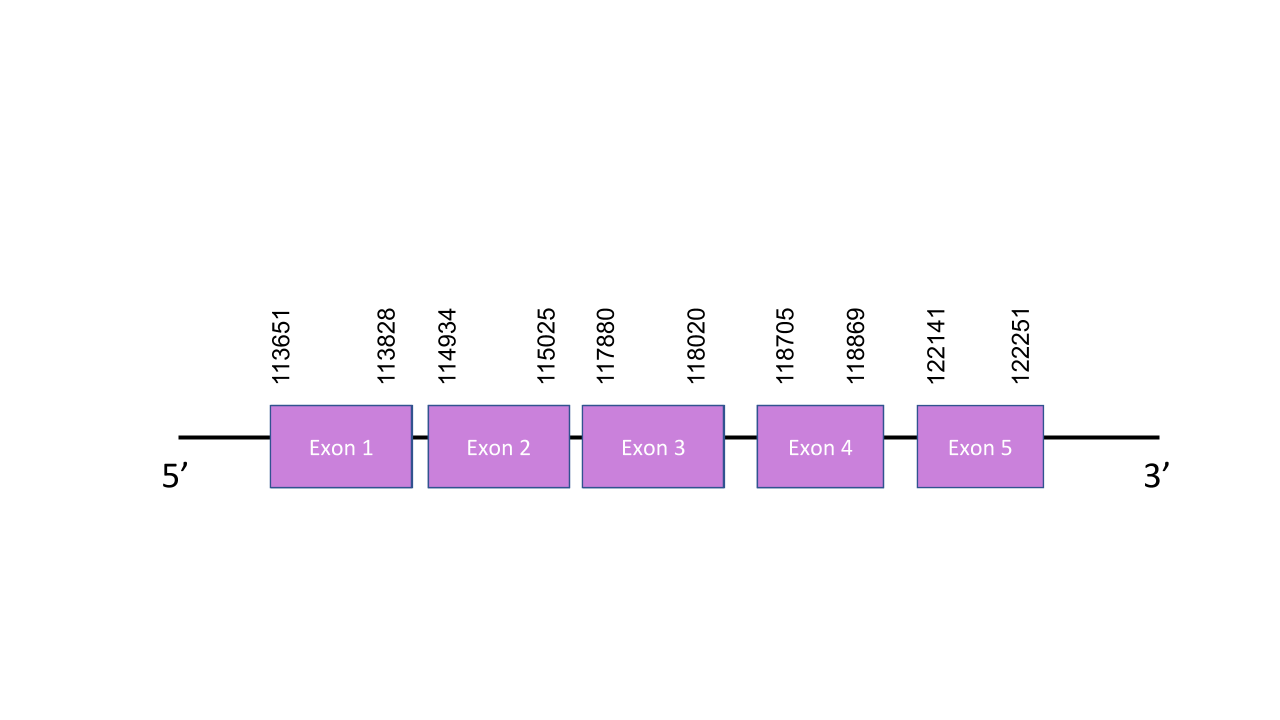
SelU2
The gene that encodes the protein SelenoU2 is located in the scaffold PISW01002714.1 between the positions 133160 and 143567 in the reverse strand. This gene has six exons as predicted with exonerate, located at 133160-133284, 136538-136669, 141598-141703, 141870-141923, 142923-142991 and 143382-143567. Seblastian was not able to predict a known selenoprotein from the Mungos mungo sequence nor any SECIS structures. After aligning the predicted protein with the human selenoprotein SelH, we obtained a perfect score. We could not find selenocysteine residues in the predicted protein, but that makes sense since as it is seen in the human homologs, it is likely that the selenocysteine residue was substituted by a cysteine in a common ancestor. Altogether, this results make it hard to tell if there is for sure SelU1 in the Mungos mungo genome. Still, the e-value, score and identity scores obtained when comparing the predicted sequence with the human one, make it seem likely that there is.
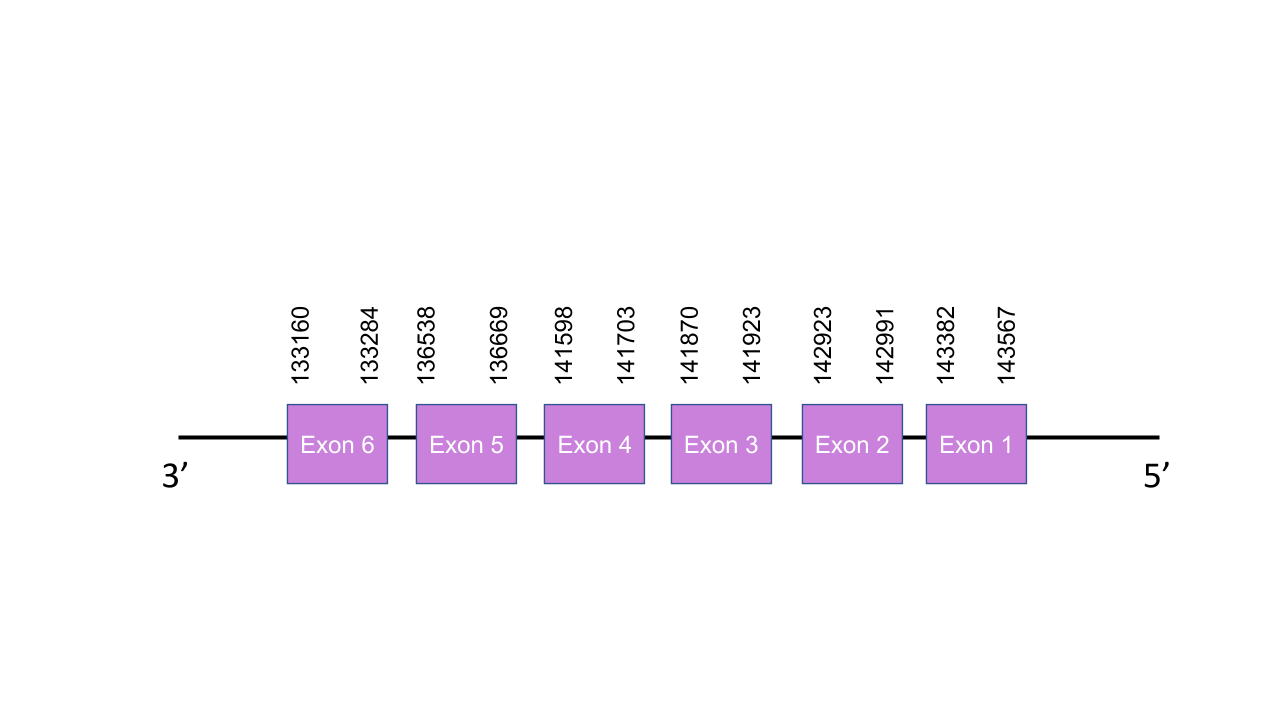
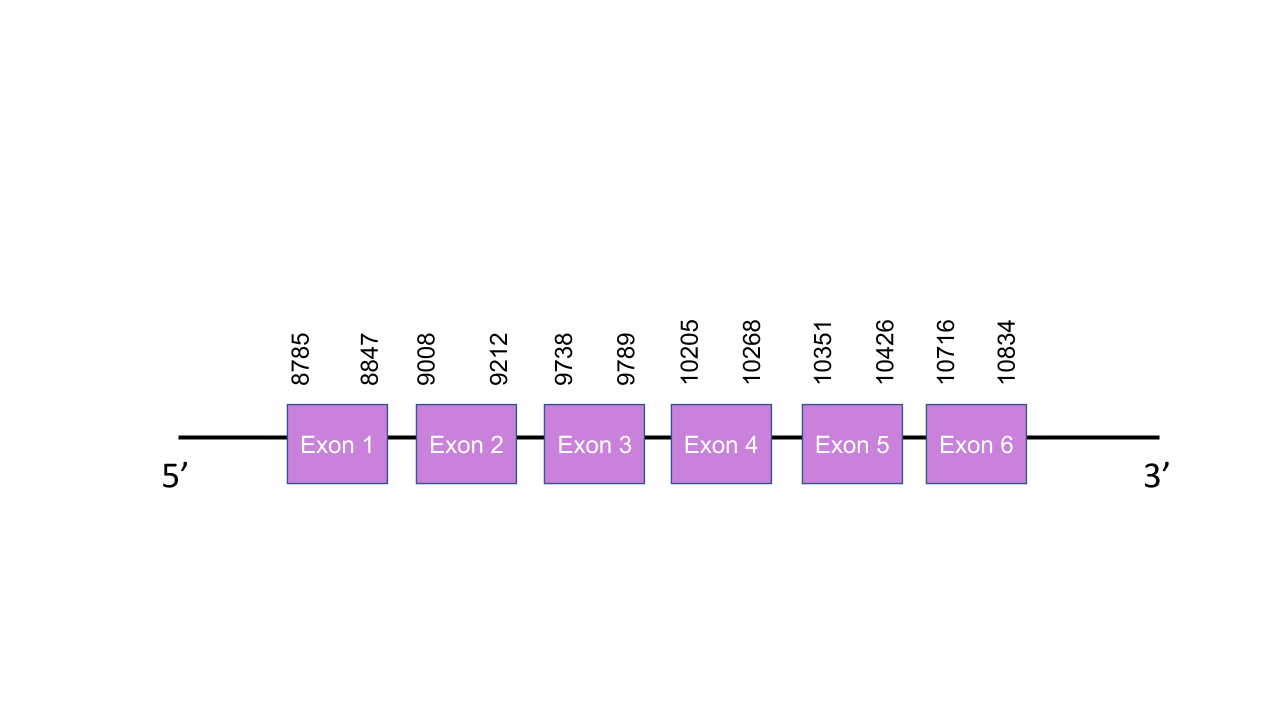
SelU3
The gene that encodes the protein SelenoU3 is located in the scaffold PISW01016784.1 between the positions 8785 and 10834 of the forward strand. This gene has six exons as predicted with exonerate, located at 8785-8847, 9008-9212, 9738-9789, 10205-10268, 10351-10426 and 10716-10834. Seblastian was not able to predict a known selenoprotein from this Mungos mungo sequence. Also, a grade B SECIS structure was found between the positions 4470 and 4554 of the forward strand -this is not a viable SECIS structure, since it is located in the same strand of our predicted gene but it is before it. After aligning the predicted protein with the human selenoprotein SelU1, we obtained a perfect score of 999/1.000. We found out that there was no selenocysteine residue in the mongoose protein because it was a cysteine homolog, still, it makes sense since the human SelU3 is as well. It looks likely that Mungos mungo encodes for the protein SelU3 i its genome, still, the lack of evidence make it hard to prove it for sure.
Machinery
Eukaryotic elongation factor (eEFsec)
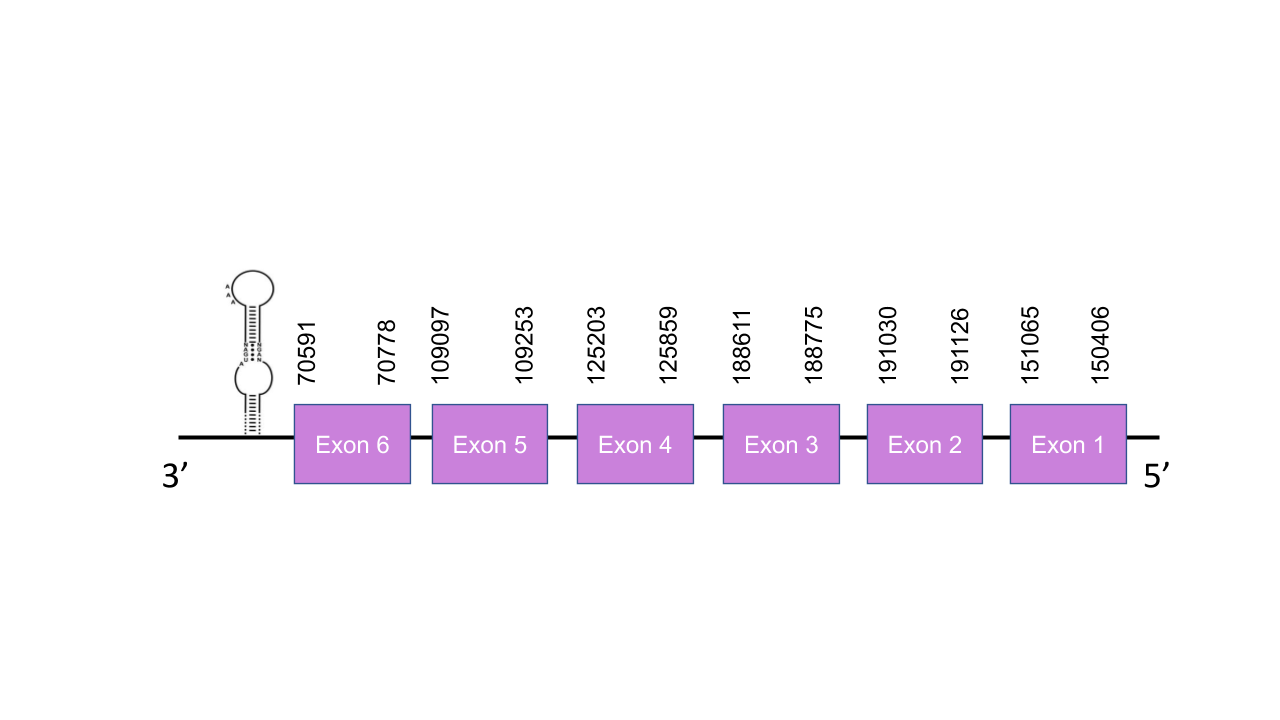
The gene that encodes the protein eEFsec is located in the scaffold PISW01000142.1 between the positions 70594-219095 in the reverse strand. Among the 4 scaffolds obtained on the Tblastn, we selected the scaffold PISW01000142.1 since it was the one with the best and highest e-value (6.68e-128) and its 6 alignment fragments has an average identity of 86,17. This gene has 6 exons predicted with exonerate, located at 151065-150406, 191030-191126, 188611-188775, 125203-125859, 109097-109253, 70591-70778, all of them in the negative strand. The T-coffee shown a score of 998, which is a very good alignment. Although the was pretty good, there was a gap of missing residues at the beggining of the sequence, which probably indicates that this part of the genome was not correctly annotated. Thus, was impossible to determine whether the predicted protein starts with a Methionine or not. Moreover, as we expected, it does not contain a Sec since it is a machinery protein and not a selenoprotein itself. In contrast, Seblastian was able to predict a grade B SECIS element in the negative strand between positions 47761-47839 on the 3’ end of the strand, but not any selenoprotein in this sequence. Even so, the score of T-coffee was good enough to afirm that eEFsec protein is a Cys-homolog conserved and well-aligned in Mungos mungo, accepting our prediction.
Phosphoseryl-tRNA kinase (PSTK)
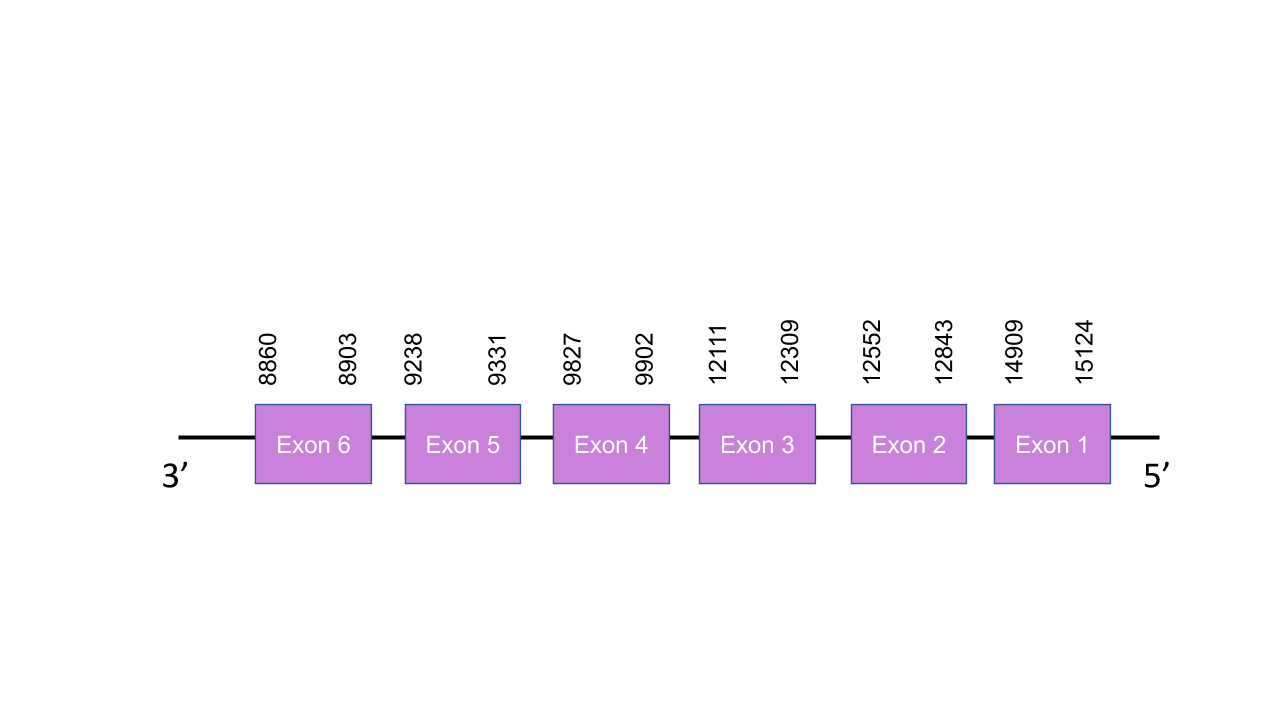
The gene that encodes the protein PSTK is located in the scaffold PISW01014100.1 between the positions 9236-15124 in the reverse strand. That was the only scaffold shown in the Tblast prediction and it has 5 fragments with a low e-value and an average identity of 83,37. This gene has 6 exons predicted with exonerate, located at 14909-15124, 12552-12843, 12111-12309, 9827-9902, 9238-9331, 8860-8903 positions, all of them in the negative strand. T-coffe shown a perfect alignment with a score of 1000. Moreover, the predicted protein starts with a Methionine. As expected, it does not present any Sec, so for being a Cys-containing homolog it presents many conserved cysteines. As expected, it has no SECIS elements neither selenoprotein predicted with Seblatian, as the Human PSTK, because it is part of the machinery and it is not a selenoprotein itself.
SECIS binding protein 2 (SBP2)
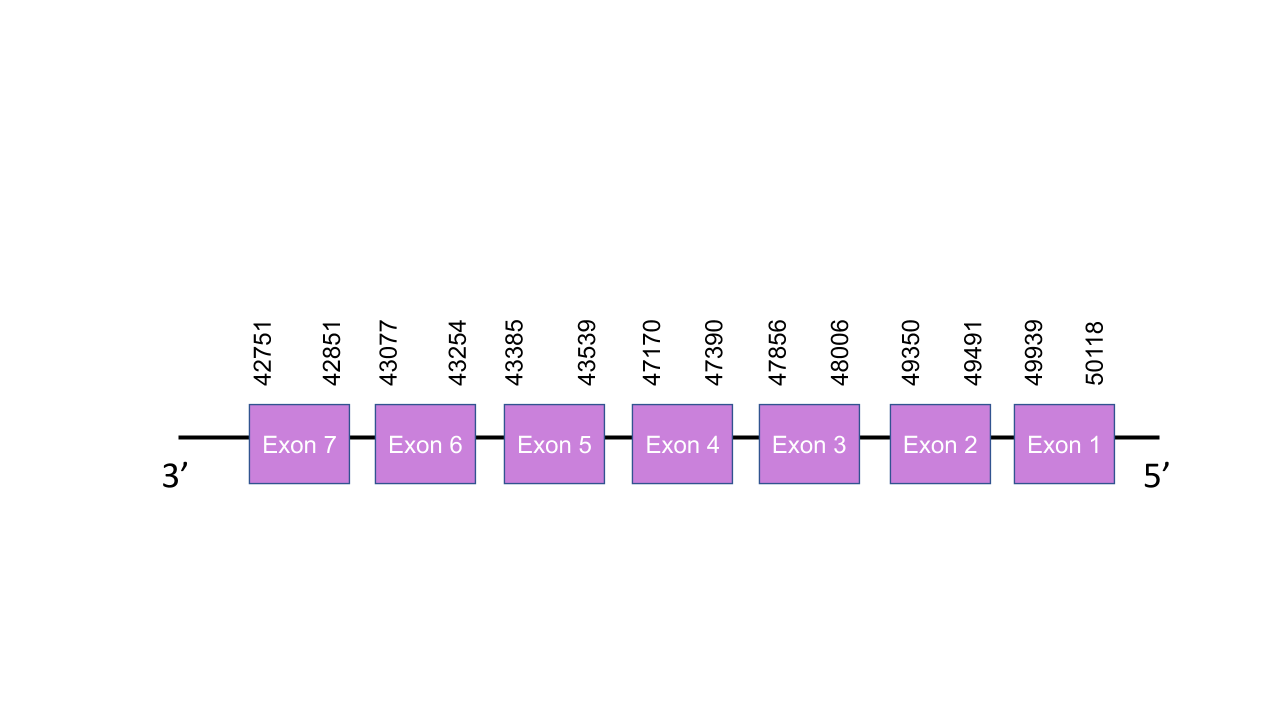
The gene that encodes the protein PSTK is located in the scaffold PISW01010000.1 between the positions 43171-50070 in the reverse strand.From the 3 different scaffols predicted with the Tblast, we selected that one since it was the one with the lowest e-value and its 5 fragments of alignment gave an average identity of 83,137. This gene has 7 exons, obtained with the exonerate prediction, located at 49939- 50118, 49350-49491, 47856-48006, 47170-47390, 43385-43539, 43077-43254, 42751-42851 positions, all of them in the negative strand. T-coffee shown a very good alignment with a socre of 986. The predicted sequence presents a huge gap at the beginning, due to a delection in the genome or a deficient annotation. Despite this, it presents many conserved cysteines as it is a Cys-containing homolog. Therefore, as we expected, neither SECIS element or selenoprotein were found by Seblastian.
tRNA Sec 1 associated protein 1 (SECp43)
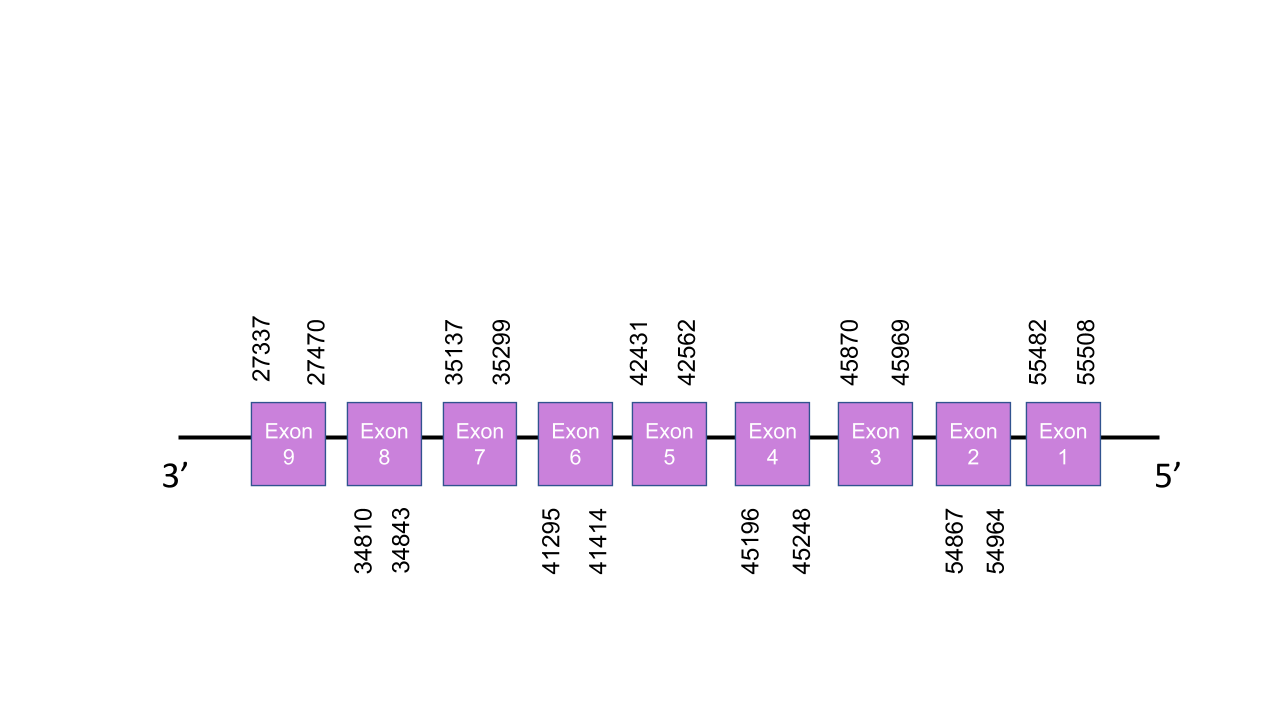
The gene that encodes the protein SECp43 is located in the scaffold PISW01002745.1 between the positions 27337-55508 in the reverse strand. Among the 12 scaffolds obtained on the Tblast prediction, we analyzed two scaffolds: PISW01004770.1 and PISW01002745.1. We finally selected the PISW01002745.1 scaffold since it was the one with the highest e-value plus an average identity of 89,88 in its 5 alignment fragments and a better T-coffee alignment with an score of 1000. This gene has 9 exons, obtained with the exonerate prediction, located at 55482-55508, 54867-54964, 45870-45969, 45196-45248, 42431-42562, 41295-41414, 35137-35299, 34810-34843 and 27337-27470 positions, all of them in the negative strand. T-coffee shown a perfect alignment with a score of 1000 between the two species. Moreover, it presents a Cys-residue as happens in the Human sequence, as we expected since it is a Cys-containing homolog. Furthermore,as we expected, since it has not a selenocysteine residue on its sequence, neither SECIS element or selenoprotein were found by Seblastian.
Selenocysteine synthase (SecS)
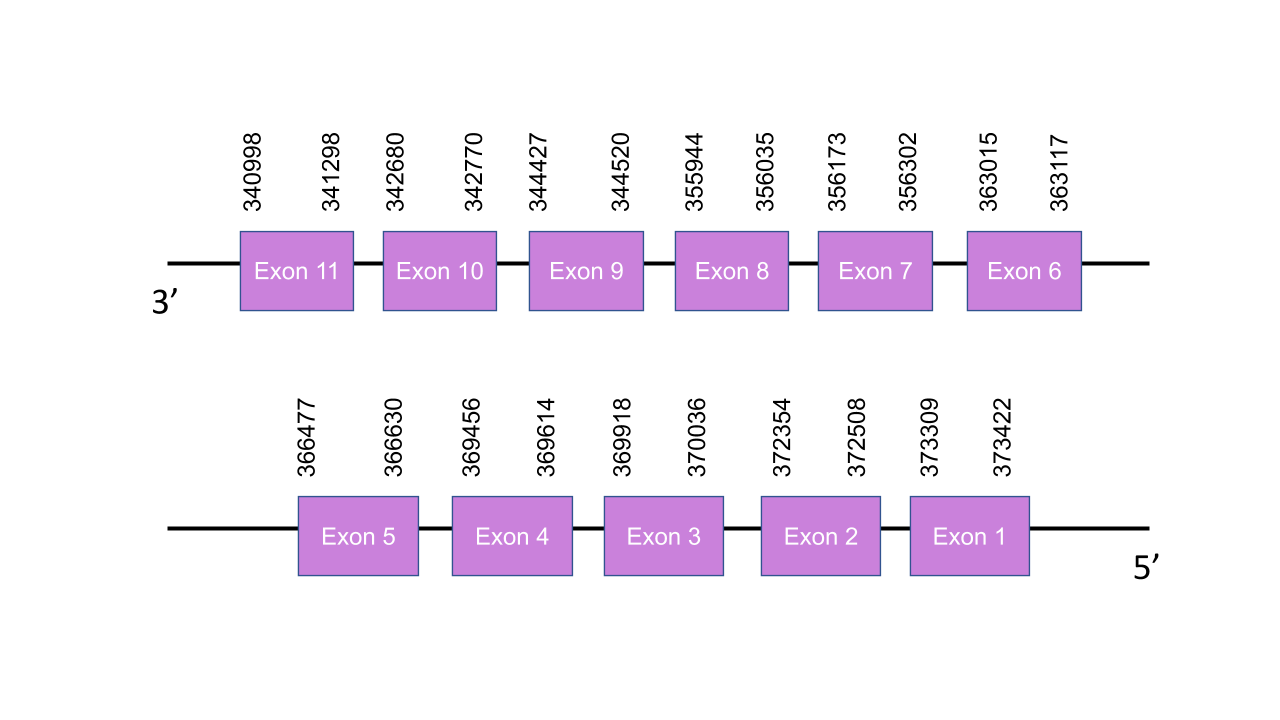
The gene that encodes the protein SecS is located in the scaffold PISW01001235.1 between the positions 340998-373422 in the reverse strand. That was the only scaffold found in the Tblast prediction and it has 11 fragments with a low e-value and an average identity of 83,37. This gene has 11 exons, obtained with the exonerate prediction, located at 340998-341298, 342680-342770, 344427-344520, 355944-356035, 356173-356302, 363015-363117, 366477-366630, 369456-369614, 369918-370036, 372354-372508, 373309-373422 positions, all of them in the negative strand. T-coffee showed an score of 998, meaning that there is a very good alignment between the two species. Moreover, the predicted protein starts with a Methionine. As expected, it does not present any Sec, so for being a Cys-containing homolog it presents many conserved cysteines. A grade B SECIS was found by Seblastian, between the positions 40446-40522. However, it was not located in the 3’ end, so this SECIS was not valid. Thus, not any valid SECIS was found. Moreover, Seblastian did not predict any selenoprotein in the sequence as expected.
Selenophosphate synthetase 1 (SPS1)
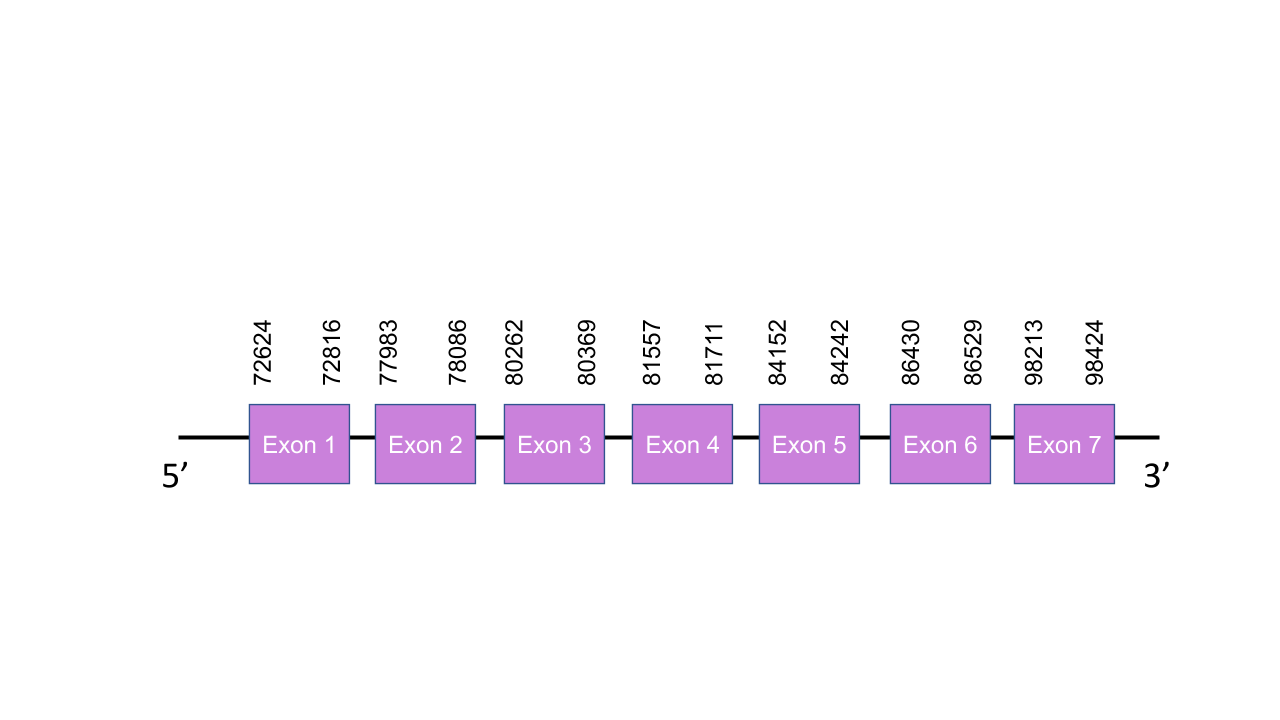
The gene that encodes the SPS1 protein is located in the scaffold PISW01006518.1 between the positions 72624-98424 in the forward strand. From the 2 different scaffols predicted with the Tblast, we selected that one since it was the one with the lowest e-value and its 2 fragments of alignment gave an average identity of 75,074 and the other one was the one defined for the SPS2. This gene contains 7 exons located at 72624-72816, 77983-78086, 80262-80369, 81557-81711, 84152-84242, 86430-86529, 98213-98424 positions of the positive strand. T-coffee showed an score of 1000, meaning that there is a perfect alignment between the two species. There was a Methionine at the start of the predicted protein. In addition, there were different align cysteines with the Cys at the human sequence, being meaningful with the fact that this protein is a Cys-containing homolog in both species. Seblastian was unable to found any SECIS element nor protein.
Selenophosphate synthetase 2 (SPS2)
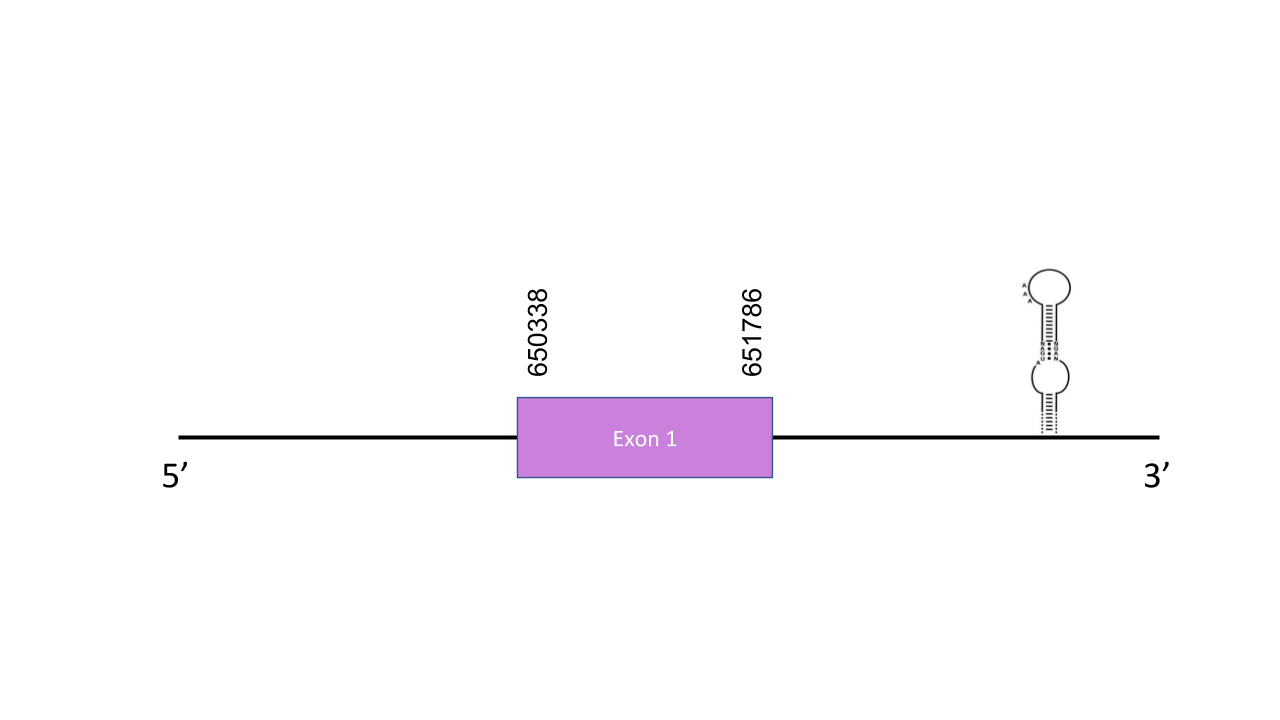
The gene that encodes the SPS2 protein is located in the scaffold PISW01000219.1 between the positions 650410-651786 at the forward strand. Tblastn prediction showed up 2 scaffolds, among them, we selected the PISW01000219.1, which has a single alignment with an average identity of 91.939 and an e-value of 0,0, meaning it is impossible this sequence was found by chance since we didn’t expect any aligment to occur just by chance for the given score. This gene has only one exon located between 650338-651786 at the positive strand. T-coffee showed an score of 997, indicating a very good alignment. The predicted protein has two Sec residues align with their human homologs. Thus, although SPS2 is a machinery protein it is a selenoprotein. Seblastian was able to predict a valid grade A SECIS element at the positions 652249-652325 of the 3’ end positive strand. Seblastian predicted a known selenoprotein as well.
Conclusions
The purpose of this assignment was to analyze and annotate the selenoproteins present in the Mungos mungo genome. We analyzed 34 selenoproteins or cysteine-homologues and 7 elements related to the selenoprotein machinery from the Homo sapiens genome in Mungos mungo. The following results were obtained:
- Selenoproteins: GPx1,GPx2,GPx3, DIO1, DIO2, DIO3, TXNRD1, TXNRD2, TXNRD3, Sel15, Sel M, Sel N, Sel O, Sel P, Sel K, Sel S, Sel H and MSRB1.
- Cysteine-containing homologues: GPx5,GPx6,GPx7,GPx8, W2, Sel T, MsrA, SelU1, SelU2, SelU3, MSRB2 and MSRB3.
- Selenoprotein machinery: eEFsec, PSTK, SBP2, SECp43, SecS, SPS1 and SPS2.
- Imprecisely predicted selenoproteins: GPx4, W1, Sel V and Sel I.
The predicted proteins DIO1, W1, Seleno V, TXRD1, TXRD2, TXRD3, GPx1, GPx5 and GPx7 do not start with a methionine residue, this indicates that the start of the protein might not be included in the prediction. For this reason, additional research needs to be conducted in order to determine the start of this Mungos mungo proteins.
Regarding the SECIS and protein predictions using Seblastian, it must be pointed out that for some proteins the program could not detect any SECIS or protein. This could be due to limitations in the sensitivity of this software.
Ultimately, it cannot be confirmed whether the variations in the predicted proteins of Mungos mungo with respect to the reference ones are due to limitations of the methods or actual evolutionary changes. Hence, although our findings contribute to the knowledge on the subject, further research must be performed.
References
(1)Gromer S, Eubel J, Lee B, Jacob J. Human selenoproteins at a glance. Cellular and Molecular Life Sciences. 2005;62(21):2414-2437.
(2)Labunskyy V, Hatfield D, Gladyshev V. Selenoproteins: Molecular Pathways and Physiological Roles. Physiological Reviews. 2014;94(3):739-777.
(3)Mariotti M, Ridge P, Zhang Y, Lobanov A, Pringle T, Guigo R et al. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE. 2012;7(3):e33066.
(4)Kasaikina M, Hatfield D, Gladyshev V. Understanding selenoprotein function and regulation through the use of rodent models. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(9):1633-1642.
(5)Metanis N, Hilvert D. Natural and synthetic selenoproteins. Current Opinion in Chemical Biology. 2014;22:27-34.
(6)Papp L, Holmgren A, Khanna K. Selenium and Selenoproteins in Health and Disease. Antioxidants & Redox Signaling. 2010;12(7):793-795.
(7)Mungos mungo (banded mongoose) [Internet]. Animal Diversity Web. 2019. Available from: https://animaldiversity.org/accounts/Mungos_mungo/
(8)Hinton, H., A. Dunn. 1967. MONGOOSES Their Natural History And Behaviour. London: Oliver And Boyd Ltd.
(9)Banded mongoose [Internet]. Oregon Zoo. 2019. Available from: http://www.oregonzoo.org/Cards/Rainforest/mongoose.banded.htm
(10)The IUCN Red List of Threatened Species [Internet]. IUCN Red List of Threatened Species. 2019 . Available from: https://www.iucnredlist.org/species/41621/45208886 .
Acknowledgements
To start with, we would like to warmly acknowledge our tutor Laura Jimenez, who selflessly aided us with the website and the many coding issues that were to come.
We were also kindly aided by Núria Centeno, who helped us in our aim to create tridimensional models of our predicted proteins. That was not possible, but it’s intention that counts.
We would also like to thank our teachers, Roderic Guigó, Robert Castelo, Cedric Notredame, Toni Gabaldón and Aida Ripoll, for challenging us with such a huge project. It got the best of us, like our self-learning capacity, and it demonstrated that we can face pretty much anything after that.
Last, but not least, we would like to thank our classmates for their invaluable help, they were the live demonstration of the quote “we lift by raising others” and they are the main reason we are here today.
Again, thanks a lot.
“Satisfaction lies in the effort, not in the attainment, full effort is full victory” - Mahatma Gandhi