The aim of the present study is the characterization and annotation of selenoproteins present in Kobus ellipsiprymnus genome. Selenoproteins are a particular type of proteins that present the amino acid selenocysteine in their catalytic redox-active site. This amino acid is encoded by a UGA codon. Ribosomes do not mistake them with UGA stop codons due to the presence of a cis-acting Sec insertion sequence (Secis element) present in the 3’UTR of the mRNA. Thanks to bioinformatics, the study and characterization of selenoproteins has been possible in many species.
An homology-based approach using H. sapiens genome has been used in order to identify selenoproteins in the given genome. Nevertheless, some proteins have been annotated using the Bos taurus genome. A program in Perl was created in order to automatize the analysis of the proteins. In this program, a protein was automatically analyzed with tblastn, exonerate, and t-coffee. As a result, an alignment between the protein of reference and the predicted protein in Kobus ellipsiprymnus was obtained. Additionally, Seblastian was used to predict possible selenoproteins and Secis elements in the giving sequence.
In this study, 22 selenoproteins, 7 cysteine-homologous proteins and 4 selenoprotein machinery proteins were identified in Kobus ellipsiprymnus genome. On the other hand, 9 selenoproteins found in H. sapiens were not predicted as selenoproteins in our animal, and one protein was not even found in Kobus ellipsiprymnus genome. All together, it is expected that the results summarized in the present article might help clarify the Kobus ellipsiprymnus genome annotation.
 There are 22 amino acids that are naturally incorporated into peptides and they are known as proteinogenic or natural amino acids. Although there are 20 amino acids encoded by the universal genetic code, two of them are encoded via variant codons and can only be incorporated into the proteins by synthetic mechanisms: selenocysteine and pyrrolysine [1,2]. In this project, we will analyze the conservation of selenoproteins in the genome of Kobus ellipsiprymnus in comparison with the ones found in the human genome.
There are 22 amino acids that are naturally incorporated into peptides and they are known as proteinogenic or natural amino acids. Although there are 20 amino acids encoded by the universal genetic code, two of them are encoded via variant codons and can only be incorporated into the proteins by synthetic mechanisms: selenocysteine and pyrrolysine [1,2]. In this project, we will analyze the conservation of selenoproteins in the genome of Kobus ellipsiprymnus in comparison with the ones found in the human genome.
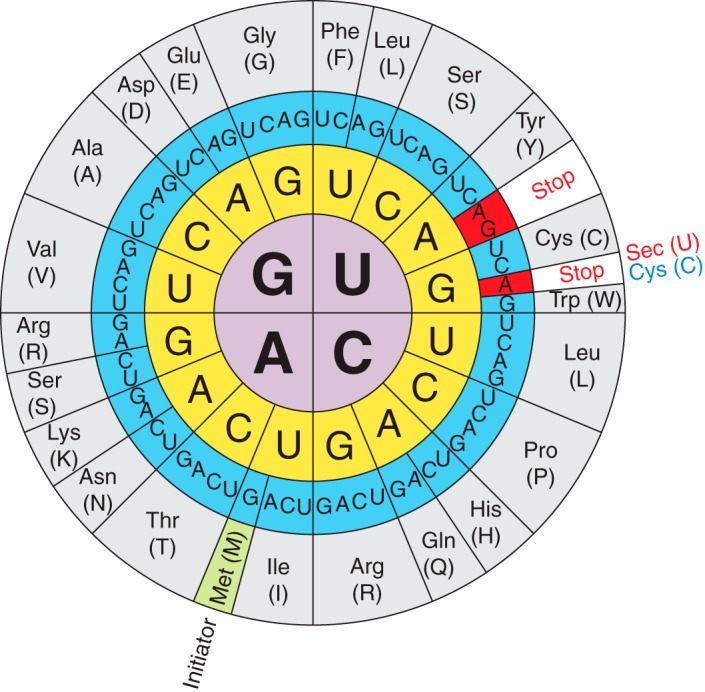
Fig 1. Selenocystein's codon in the genetic code.
During many years selenium (Se) had been considered a toxic element. However, in the past 50 years the perspective has changed and it is now classified as an essential micronutrient in life that has several important health benefits [1]. Se deficiency is associated to numerous pathophysiological conditions, such as cancer, neuromuscular disorders, heart disease or inflammation. Moreover, it also contributes in mammalian development, immune function and inhibition of viral expression [2,3]. Since one of the major forms of Se in the cell is being part of selenocysteines (Sec, U), this amino acid and selenocysteine-containing proteins are being deeply studied [2].
Selenocysteine is considered as the 21st amino acid and the micronutrient. Sec structure is nearly identical with cysteine’s (Cys) one, but it contains Se instead of sulfur (S), which confers an enhanced catalytic activity to proteins that contains the Sec [2,3,4].
Fig 2. Comparison between the structure of the amino acids selenocysteine and cysteine. 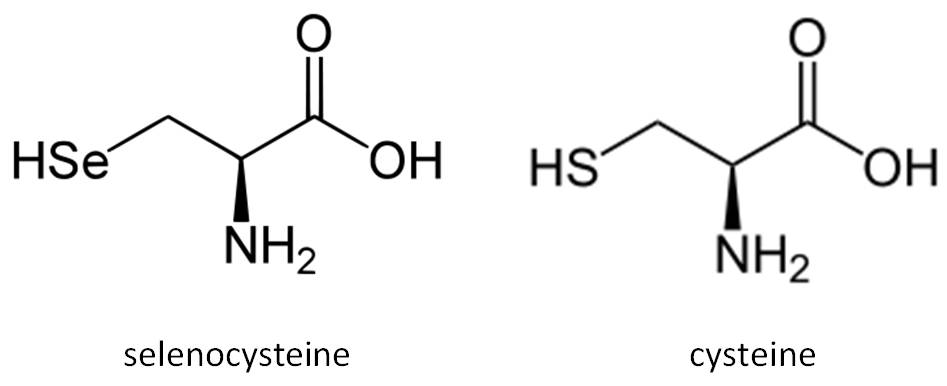
Once Sec is synthesized, it is inserted into selenocysteine-containing protein, known as selenoproteins [5]. Up to now, 25 human genes have been identified as selenoprotein encoders. Through the study of the selenoproteins in humans, the human selenoproteome has been established [2]. Some of the best known families are Glutathione peroxidase, thioredoxin reductases, methionine sulfoxide reductase and selenoproteins located in the endoplasmic reticulum [4].
Selenoproteins are enzymes with oxidoreductase function which have the Sec in the catalytic redox active site [6]. Therefore, they have important functions in the cell, such as antioxidant defense, cell signaling and redox homeostasis [4]. The redox homeostasis function is conserved among many species. Interestingly, selenoproteins perform the same activities as their homologues - proteins that contain Cys instead of Sec - but in a more efficient manner [6].
Sec is encoded by a UGA codon in prokaryotes, archaebacteria and eukaryotes. The UGA codon is mainly a stop codon, indicating the termination of protein synthesis. Nonetheless, ribosomes do not confuse the UGA stop codon and the UGA Sec-coding codon. There are two main aspects that make it possible. First of all, Sec is the only amino acid in eukaryotes which is synthesized in its own tRNA, called Sec-tRNA[Ser]Sec. Second, the decoding of UGA Sec-coding codon depends on cis-acting sequences present in the selenoprotein mRNA, known as cis-acting Sec insertion sequence (SECIS) [1,2].
Sec synthesis starts when the conventional seryl-tRNA synthetase charges a serine on the tRNASec, becoming a Ser-tRNASec. Serine is then phosphorylated to phosphoserine (Sep) by the O-phosphoseryl-tRNASec kinase (PSTK). Afterwards, the selenocysteine synthase SecS catalyzes the conversion of Sec-tRNASec to Sec-tRNA[Ser]Sec. This enzyme uses monoselenophosphate as a donner. The enzyme which produces the monoselenophosphate is a selenoprotein itself, as it has a Sec in the active site, and it is known as selenophosphate synthetase (SPS2) [1,9].
Once the Sec-tRNA[Ser]Sec has been synthetized, it has to be incorporated in the selenoprotein. In order to do that, some elements have an important role during the translation of the mRNA: SECIS element and trans-acting proteins. SECIS elements are defined as are cis-acting stem-loop RNA structures that are found in the 3′-untranslated regions (3’ UTR) of all eukaryotic and archaeal selenoprotein mRNAs [2]. They can be is located in quite far from the UGA/Sec codon, even over 4000 nt far from it. Even though SECIS are not highly conserved regarding their nucleotide sequence, all of them have a similar stem-loop structure. Their structure is important because it is thought to be the place where protein factors required of the Sec incorporation bind [1].
Instead of the general elongation factor eFA1A, the Specialized Translation Elongation Factor EFSec is required for the decoding of the UGA/Sec. EFSec specifically binds Sec-tRNA[Ser]Sec, but not Sec-tRNASec, allowing the incorporation of this tRNA to the protein. It also has a domain that allows the interaction with SBP2 [9].
SECIS-Binding protein 2 (SBP2) is RNA-binding protein found that binds to both SECIS elements and ribosomes. As mentioned before, it binds to the EFSec, which recruits Sec-tRNA[Ser]Sec [1,9].
The ribosomal protein L30 competes efficiently with SBP2 for binding to SECIS element. Therefore, it is thought to displaces transiently SBP2 in order to place the SECIS closer to the ribosomal A site [9].
When a ribosome reaches a UGA codon in presence of a SECIS element, the previously mentioned proteins act. Consequently, the ribosome will interact with the EFSec and will incorporate the Sec-tRNA[Ser]Sec. The presence of SBP2 and L30 are also required in this process [1,9].
Fig 3. Schematic representation of selenoproteins' synthesis.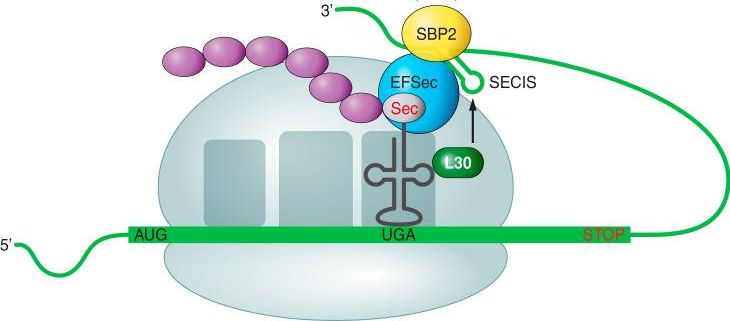
Even though selenoproteins are not present in all the species, they have been identified in the three domains of life -bacteria, archaea and eukaryotes-, as well as in viruses [2,6]. In addition, some of the selenoprotein families partially overlap in both prokaryotic and eukaryotic cells [6,7].
Genomes of several vertebrates have been analyzed in order to study Sec- and Cys-containing homologs proteins, and results reaffirm that the mammalian selenoproteome has remained relatively stable [8]. Bioinformatics has allowed the characterization of the origin and loss of each selenoprotein from fish to mammals and, through these analysis, it is known that the ancestral selenoproteome included 28 selenoproteins [8].
Since this ancestral selenoproteome, duplications and deletions of genes encoding for selenoproteins have occurred. Therefore, eukaryotes have a highly variable number of selenoproteins: from zero in higher plants, to more than 40 in some fishes [7]. This variation, is considered to be interesting in order to study how environmental and other factors have influence in the the evolution of eukaryotic selenoproteome [7].
| Kingdom | Phylum | Class | Order |
|---|---|---|---|
| Animalia | Chordata | Mammalia | Cetartiodactyla |
| Family | Subfamily | Genus | Specie |
|---|---|---|---|
| Bovidae | Reduncinae | Kobus | K. ellipsiprymnus |
Fig 4. K. ellipsiprymnus. 
Etymologically, the name K. ellipsiprymnus (Ogilbyi, 1833) comes from greek, ellipes (ellipse) and prymnos (postern part) due to the presence of a white ring on the rump. Their vernacular name is waterbuck as a result of their high dependence with water [10].
Thirteen subspecies have been identified and classified into two groups, based on their characteristic rump patterns, fur colour and geographic distribution. The two groups are the following ones: common (Kobus ellipsiprymnus ellipsiprymnus) and defassa (Kobus ellipsiprymnus defassa).
The ellipsen waterbuck group (K. e. ellipsiprymnus) is located throughout most of sub-Saharan Africa, whereas the defassa waterbuck group (K. e. defassa) is found west of the Gregory Rift and south of the Sahel from Eritrea to Guinea Bissau. Distribution of both groups slightly overlaps along the Rift Valley in Kenya and Tanzania [11].
Fig 5. Distribution of subspecies of Kobus ellipsiprymnus.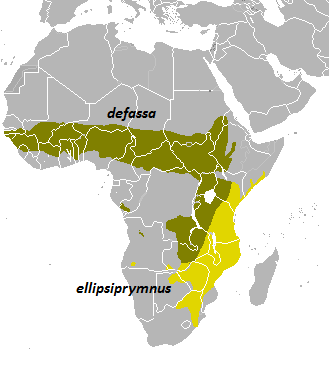
Weight: Males: 198-262 kg, Females: 161-214 kg.
Body length: 180-220 cm.
Shoulder length: 100-130 cm.
Kobus ellipsiprymnus is one of the largest antelopes amongst the species of Kobus. It is sexually dimorphic. More specifically, growth is faster and weight is bigger in males than in females. Their color varies between auburn and grey.
Moreover, common waterbuck, K. e. ellipsiprymnus has a distinctive white ring around the rump, whereas the defassa waterbuck, K. e. defassa, doesn’t have it [12].
Fig 6. K. e. defassa (left side) and K. e. ellipsiprymnus (right side).
 Sexual maturity: Males: 6 years, females: 3 years.
Sexual maturity: Males: 6 years, females: 3 years.
Gestation period: 9 months.
Average number of offspring: 1.
Lifespan: Wild: 18 years, captivity: 30 years.
Most births occur during the wetter seasons (August and November). After birth, newborns are isolated in thickets away from the herd. This isolation takes place from two or three weeks-old up to two months. Calves are weaned when they are eight months-old. At this moment, horns begin to form on males and then, they join groups of calves of their own age [13].
Fig 7. Defassa waterbuck adult and offspring.
As its name suggests, the waterbuck is a good swimmer and it flees into water if it is pursued. However, it has been reported that they do not actually like going into water.
Generally, waterbuck prefer savanna, grassland and scrub habitats that are close to water for feeding. Grass, leaves and reeds constitute a substantial 70-95% of their diet. It has been studied that during the dry season about 32% of their 24-hours day is spent on browsing, whereas they do not spend time performing this activity during the wet season [14].
Wild waterbuck live in well divided ranges which are made up of 30 individuals, many of them are females and there also territorial and non-territorial males. Upon maturation, when they are around 6-years old, male waterbuck becomes territorial and only 5-10% of mature territorial males can live together.
The herd has a social hierarchy based on size and strength, and contests are frequent.
Waterbucks are chased, hunted and poached for food and sport as they are specifically susceptible due to their sedentary nature. As a result, waterbucks population’s current trend is to decrease. According to IUCN red list criteria, defassa waterbuck group is classified as “near threatened” specie and common waterbuck group is listed as a “least concern” specie [14]. Most of Kobus ellipsiprymnus population survive in protected areas.
In order to characterize the Kobus ellipsiprymnus’ selenoproteins, an homology-based study was performed. The selenoproteins used as a query came from Homo sapiens and were extracted from the database SelenoDB 1.0. The proteins were obtained from SelenoDB 1.0 as this version of the database is more accurate than the latest version. For those selenoproteins in which the predicted protein in Kobus ellipsiprymnus was not as good as expected (i.e. a bad alignment in t-coffee, the prediction of only one part of the protein, a bad score in t-coffee) the selenoprotein from Bos taurus (cow) was used. Bos taurus’ selenoproteins were extracted from SelenoDB 2.0, as this animal is not present in SelenoDB 1.0. The selenoproteins of Bos taurus were used as a query because both B.taurus and K. ellipsiprymnus are classified in the family of Bovidae, being B.taurus closer to K. ellipsiprymnus than H.sapiens.
The proteins used as a query are shown in the following table:
| Origin | Selenoproteins |
|---|---|
| Homo sapiens | GPx1, GPx2, GPx3, GPx4, GPx7, GPx8, DI1, DI2, DI3,SBP2, SPS1, Sel15, SelI, SelK, SelN, SelO, SelR1, SelR2, eEFSec, SelS, SelT, SelU1, SelU2, SelU3, SelW1, SelW2, TR1, TR2, TR3 |
| Homo sapiens + Bos taurus | GPx5, GPx6, MsrA, SelH, SelM, SelP, SelR3, SelV |
Kobus ellipsiprymnus’ genome has been given to us. We will use the following path in order to access:
Kobus ellipsiprymnus’index has also been given to us:
Afterwards, a program in Perl was created in order to automate the whole process of analyzing the proteins. First of all, each sequence was saved in a different position of a vector. By going through all the positions of the vector, and therefore, all the queried Selenoproteins, the homology to the given genome will be checked. The program can be found here. TBLASTN Tblastn will search for the sequence of the given Selenoprotein in the genome of interest. Once run, it will tell us the number of hits, the length of the hit (starting point and ending point), the corresponding e-values, the scaffold of the hit, and the percentage of coincidence. FASTAFETCH FASTSUBSEQ EXONERATE
Once the prediction of the exons embedded in the comprised region is done, egrep will extract them and save them all together in a variable. FASTASEQFROMGFF FASTA TRANSLATE T-COFFEE SEBLASTIAN and SECIS Search
In order to run all the programs correctly, selenocysteines were replaced by “X” at the beginning of the analysis (tblastn does not recognize selenocysteines). In the same way, the symbols # and % present in the FASTA sequence of selenoproteins were removed automatically.
Once the iteration has begun (the length of the vector determines the number of loops, and therefore the number of Selenoproteins that we are going to check for homology), the program will run tblastn, given a specific Selenoprotein FASTA file and the genome of interest.
It will be printed a summary of the tblastn results filtering only those hits which have e-values smaller than 0,0001 (in other words, those of them which are significant).
The program will then ask the user to introduce the name of the scaffold with the smallest e-value and the corresponding starting point of the hit. The length for each particular hit has been automated as we know both the starting point of the hit and the length of the scaffold.
$inici=
chomp ($inici);
$inici=$inici - 100000;
if ($inici<0)
{
$inici=0;
}
open (my $FILE,"<", "/home/u137251/Desktop/gen_len.txt") or die "Can't open\n";
while(my $row =<$FILE>){
chomp $row;
my @linea = split(/\t/, $row);
$hash{$linea[1]}=$linea[0];
}
close($FILE);
if (($inici+$length)>$hash{$nomdelhit}){
$length=$hash{$nomdelhit};
$length = $length - $inici -1;
}
Afterwards, the program will run fastafetch (one of many exonerate programs). Fastafetch will search for the scaffold in the genome.index file.
In order to extract a more comprised region in which the hit is held, we will use fastasubseq. Fastasubseq uses both the outcome of the fastafetch command (which has placed the hit in the genome.index file), and the values of the length and starting point of the hit (previously introduced by the user and corrected, if so, by the program).
Within the comprised region in which the hit is held, there are both introns and exons. Exonerate will predict the number and location of the exons of the predicted protein.
Afterwards, Fastaseqfromgff extracts the exons of the predicted protein creating a new file with all the exons attached (cDNA).
From the cDNA sequence of the predicted protein in Kobus ellipsiprymnus, Fasta Translate will be used in order to obtain the predicted sequence of the protein. Here, the first open reading frame will be selected by the argument -F 1.
The final step of the program is T-coffee, which enables the creation of the global alignment between two given sequences: the human or cow selenoprotein and the predicted Kobus ellipsiprymnus selenoprotein. Before performing the t-coffee, the program has to change the stop codons to “X” present in the predicted protein.
system ("/mnt/NFS_UPF/soft/tcoffee/bin/t_coffee /home/u137251/Desktop/$protein[$i] $protpred");
The Selenoprotein Prediction Server is used in this study in order to predict selenoproteins and SECIS elements given a specific sequence. This sequence is extracted from fastasubseq (and it contains the nucleotide sequence in which the selected hit with the lowest e-value and best tcoffee score was placed).
This program has a main restriction: the input file has to be maximum 100.000 nucleotides. As explained before, in the fastasubseq program, it was chosen a length of 200.000, so the file obtained before for each program can not be used here. In order to solve this problem, another Perl program was performed. This program takes the tblastn previously run for each protein. Afterwards, the fastasubseq is specially programmed to use a length of 100.000 nucleotides taking into account in which strand is located the hit and the fact that the SECIS element is found in the 3’ region of the protein.
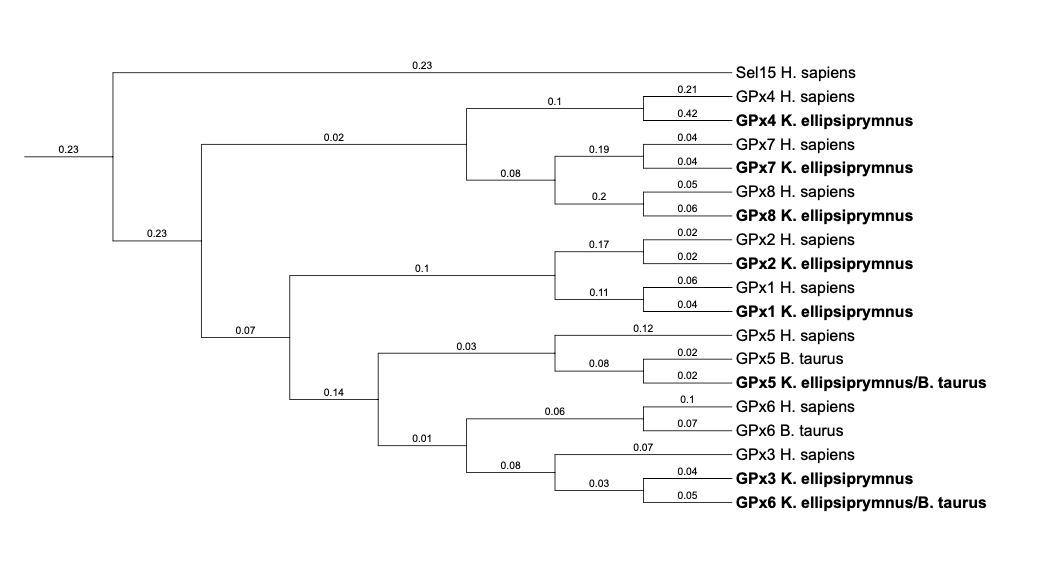
Finally, phylogenetic trees were done with the different predicted proteins and the proteins taken as a reference. It was done using Clustal Omega and iTOL.
The results obtained when predicting selenoproteins on Kobus ellipsiprymnus genome looking for homologous of human selenoproteins are exposed extensively below. Each selenoprotein is discussed individually. When the predicted proteins obtained from the human selenoproteins were not as good as it would have been expected (not all the protein has been correctly predicted, there were gaps inside the sequence or there was a bad alignment,...), a second attempt of prediction was performed using the Bos taurus’ selenoprotein.
We can call selenoprotein a given protein only if it has both a SECIS element and at least one Selenocysteine. On the other hand, we will talk about a cysteine homologous if the given protein has conserved the SECIS element but instead of having a Selenocysteine presents a cysteine amino acid. Finally, a third possible scenario would be that no homologous exist on the genome of the Kobus ellipsiprymnus, having been lost during evolution. This third scenario will be named non-predicted selenoprotein.
The main results are summarized in the following table. In addition, the files obtained when performing the analysis are also presented here.
The correspondence between the icons and the meanings is the following:
| Icon | Meaning |
|---|---|
 |
Human protein |
 |
Bos taurus protein |
 |
Correctly predicted |
 |
Not correctly predicted |
 |
Non-existing document |
| Name | Protein sequence | Residue | tBLASTn | Scaffold | Exons | Exonerate | Gene Location | Predicted protein | T-Coffee | Seblastian | SECIS sequence | SECIs image |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPx | ||||||||||||
| GPx1 |  |
Sec |  |
SJYR01000014.1 |  |
 |
1.042.563-1.043.408 (+) |  |
 |
 |
 |
 |
| GPx2 |  |
Sec |  |
SJYR01000393.1 |  |
 |
516.903-519.965 (+) |  |
 |
 |
 |
 |
| GPx3 |  |
Sec |  |
SJYR01000531.1 |  |
 |
471.838-479.114 (+) |  |
 |
 |
 |
 |
| GPx4 |  |
Sec |  |
SJYR01000491.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01000970.1 |  |
 |
1.298.264-1.300.336 (+) |  |
 |
 |
 |
 |
|
| GPx5 |  |
Cys |  |
SJYR01000174.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Cys |  |
SJYR01000014.1 |  |
 |
- |  |
 |
 |
 |
 |
|
 |
- |  |
SJYR01000174.1 |  |
 |
488.423-516.694 (-) |  |
 |
 |
 |
 |
|
| GPx6 |  |
Cys |  |
SJYR01000174.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Cys |  |
SJYR01000174.1 |  |
 |
- |  |
 |
 |
 |
 |
|
 |
Sec |  |
SJYR01000174.1 |  |
 |
- |  |
 |
 |
 |
 |
|
 |
Sec |  |
SJYR01000531.1 |  |
 |
471.344-488.426 (+) |  |
 |
 |
 |
 |
|
| GPx7 |  |
Cys |  |
SJYR01000129.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Cys |  |
SJYR01000065.1 |  |
 |
516.812-525.517 (+) |  |
 |
 |
 |
 |
|
| GPx8 |  |
Cys |  |
SJYR01000057.1 |  |
 |
1.105.599-1.101.599 (-) |  |
 |
 |
 |
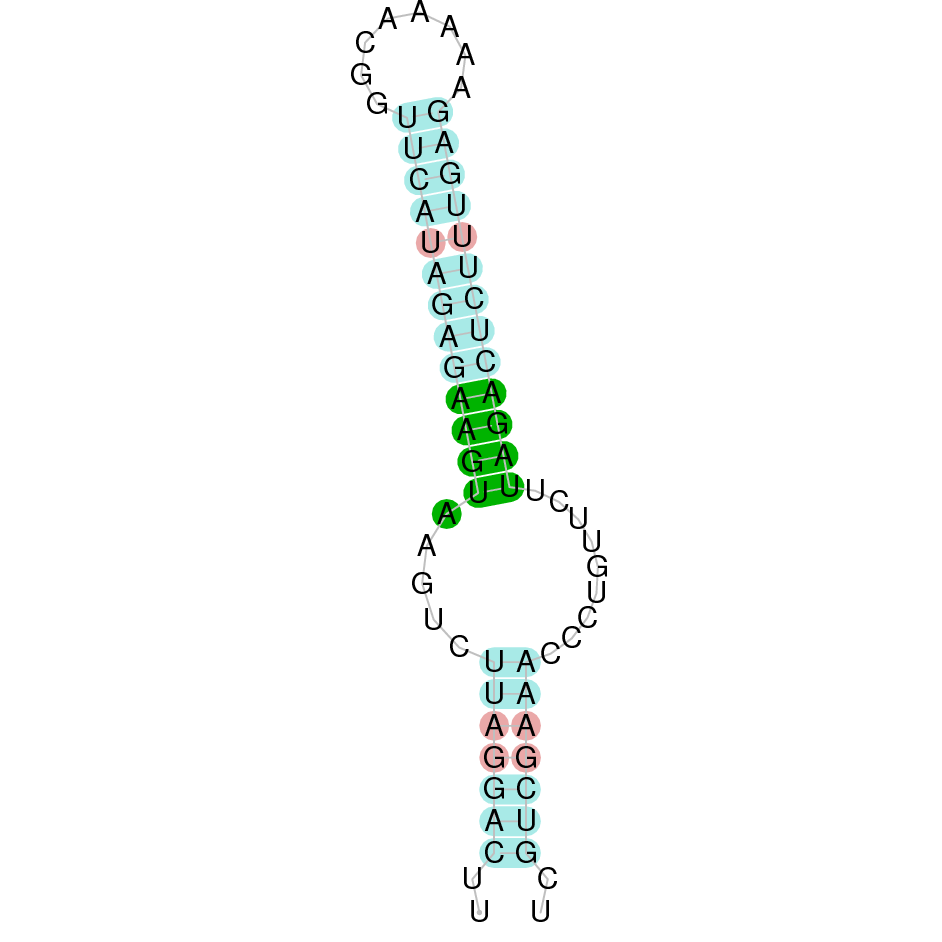 |
| DI | ||||||||||||
| DI1 |  |
Sec |  |
SJYR01000065.1 |  |
 |
1.995.803-2.013.560 (+) |  |
 |
 |
 |
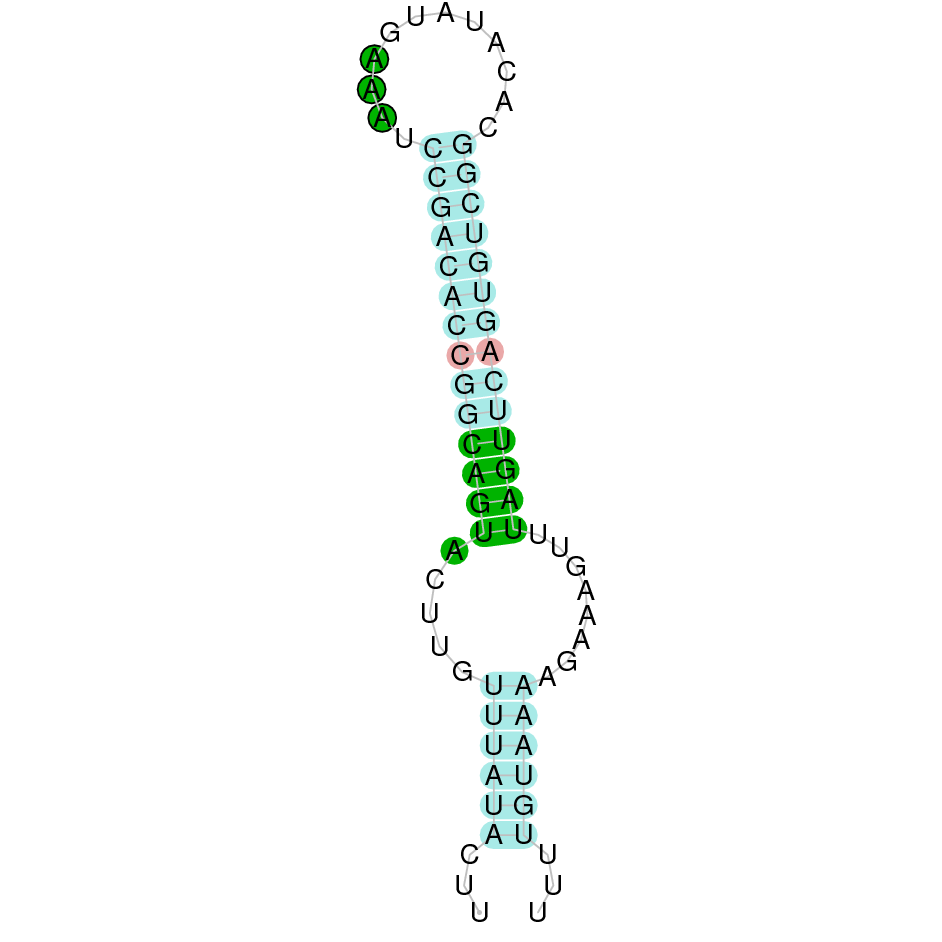 |
| DI2 |  |
Sec |  |
SJYR01000770.1 |  |
 |
734.643-744.098 (+) |  |
 |
 |
 |
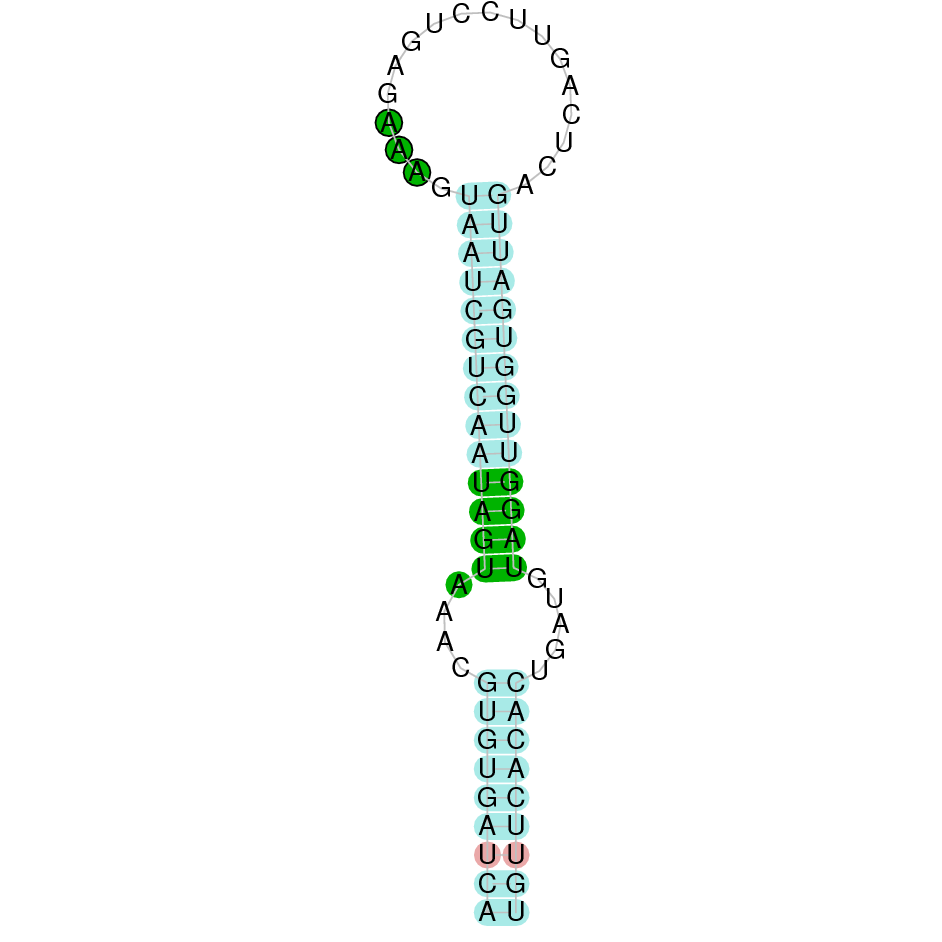 |
| DI3 |  |
Sec |  |
SJYR01002697.1 |  |
 |
103.529-104.221 (-) |  |
 |
 |
 |
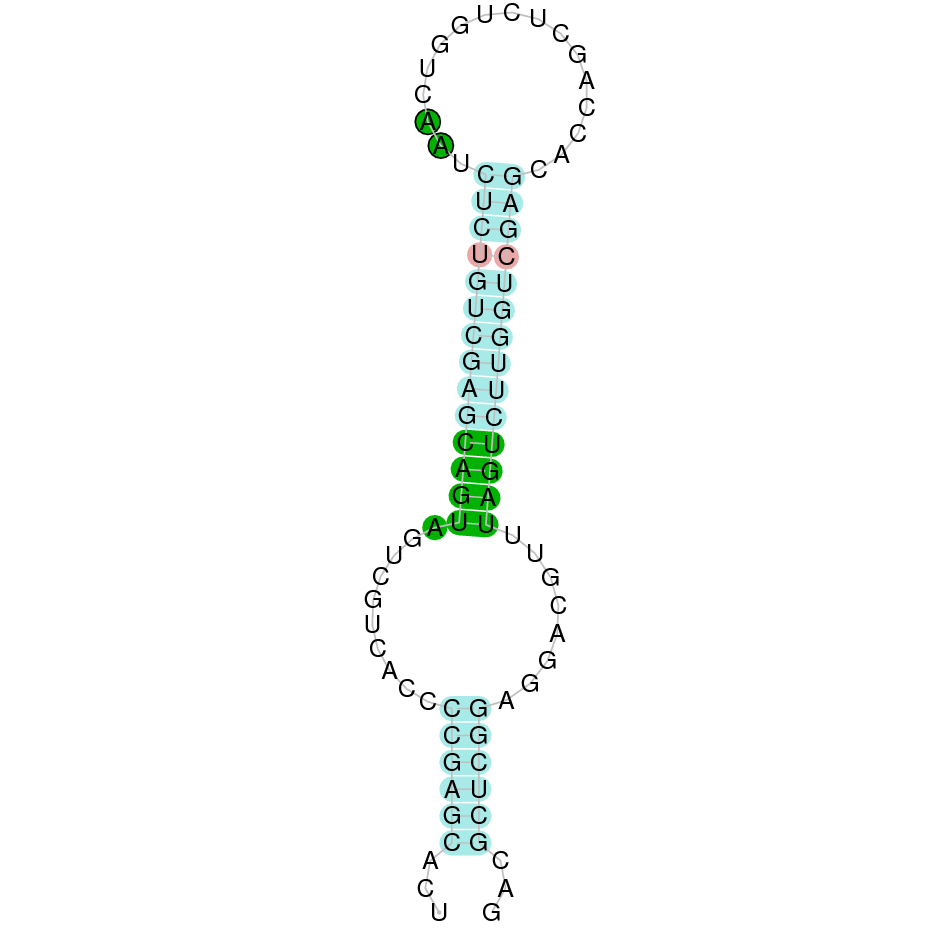 |
| MsrA | ||||||||||||
| MsrA |  |
Cys |  |
SJYR01026337.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Cys |  |
SJYR01000661.1 |  |
 |
916.411-932.031 (-) |  |
 |
 |
 |
 |
|
 |
- |  |
SJYR01026337.1 |  |
 |
- |  |
 |
 |
 |
 |
|
| Sel15 | ||||||||||||
| Sel15 |  |
Sec |  |
SJYR01001197.1 |  |
 |
16.679-56.387 (-) |  |
 |
 |
 |
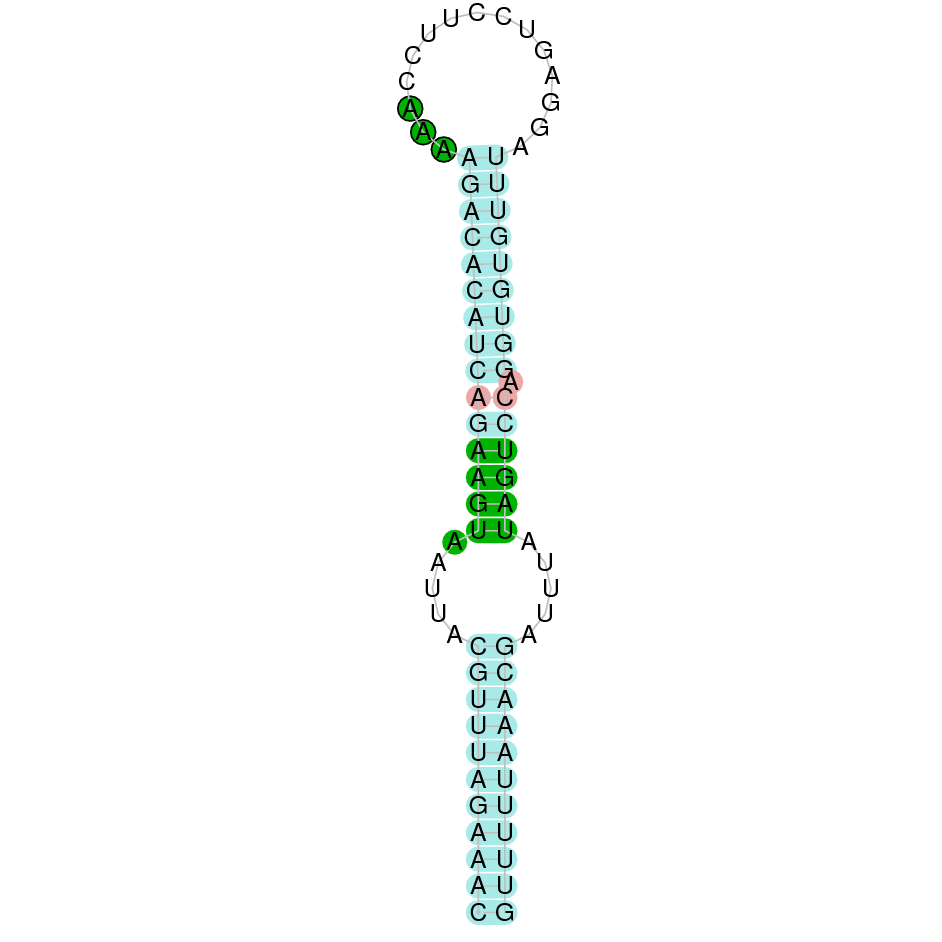 |
| SelH | ||||||||||||
| SelH |  |
Sec |  |
SJYR01002415.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01002415.1 |  |
 |
99.565-10.1372 (+) |  |
 |
 |
 |
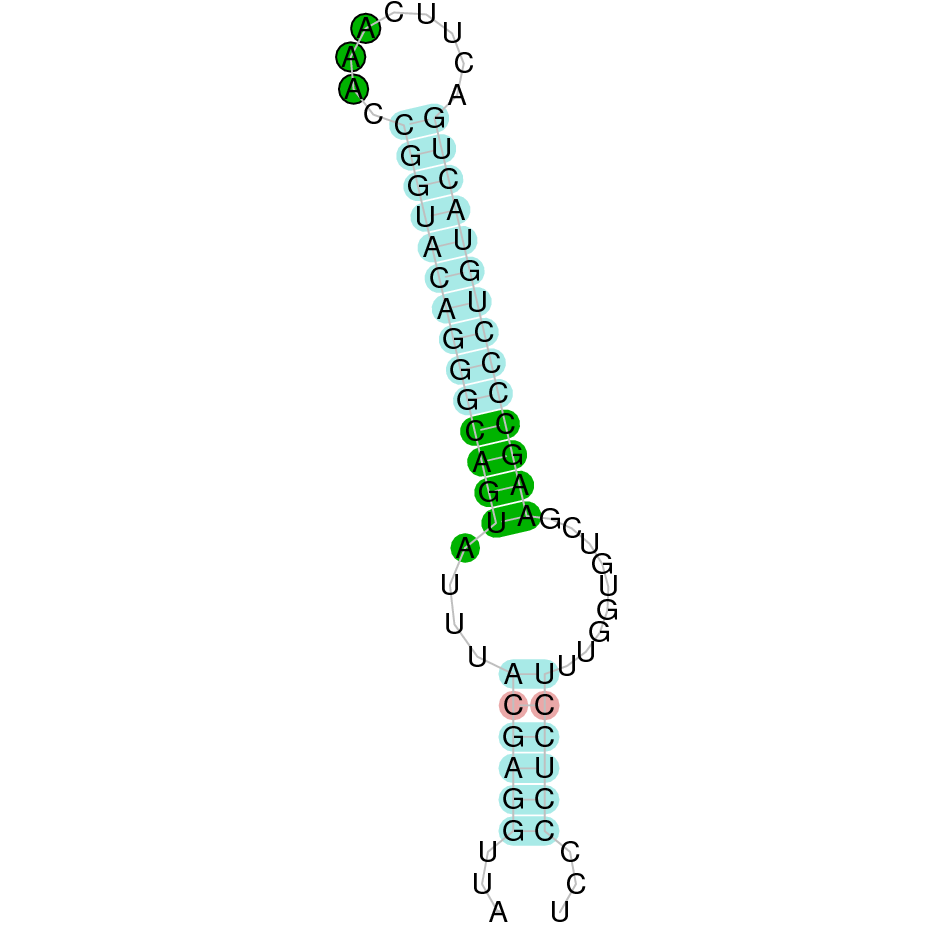 |
|
| SelT | ||||||||||||
| SelT |  |
Sec |  |
SJYR01000122.1 |  |
 |
1.600.835-1.626.204 (-) |  |
 |
 |
 |
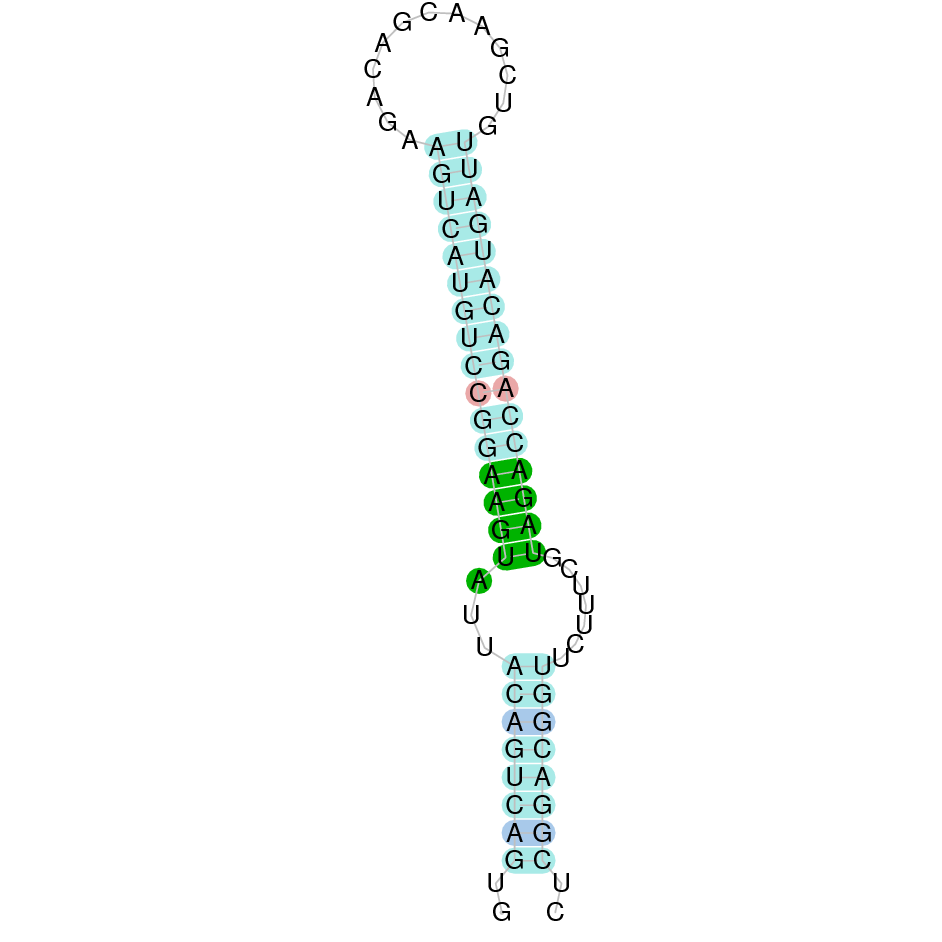 |
| SelV | ||||||||||||
| SelV |  |
Sec |  |
SJYR01001274.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01026337.1 |  |
 |
- |  |
 |
 |
 |
 |
|
| SelW | ||||||||||||
| SelW1 |  |
Sec |  |
SJYR01001274.1 |  |
 |
539.994-540.248 (+) |  |
 |
 |
 |
 |
| SelW2 |  |
Cys |  |
SJYR01003240.1 |  |
 |
145.809-146.774 (+) |  |
 |
 |
 |
 |
| SelI | ||||||||||||
| SelI |  |
Sec |  |
SJYR01000772.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01000634.1 |  |
 |
460.203-499.296 (-) |  |
 |
 |
 |
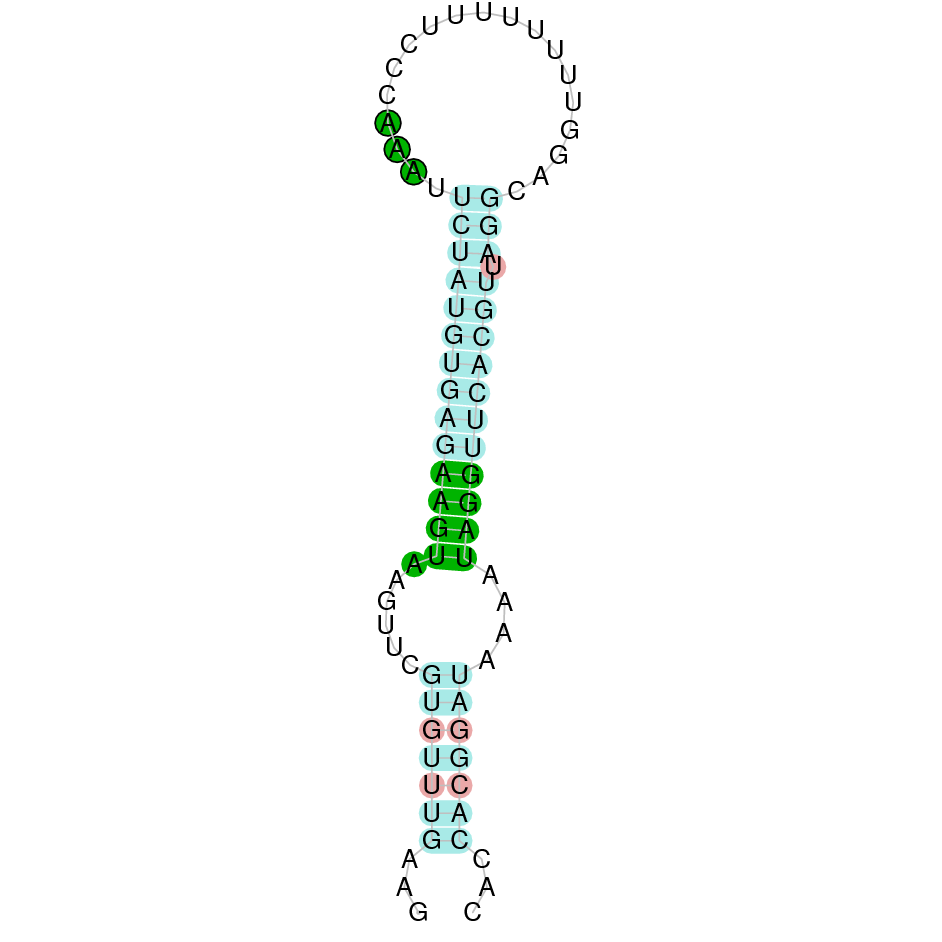 |
|
| SelK | ||||||||||||
| SelK |  |
Sec |  |
SJYR01000223.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01000121.1 |  |
 |
1.585.102-1.590.598 (-) |  |
 |
 |
 |
 |
|
| SelM | ||||||||||||
| SelM |  |
Sec |  |
SJYR01000283.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01000283.1 |  |
 |
747.033-749.501 (-) |  |
 |
 |
 |
 |
|
| SelN | ||||||||||||
| SelN |  |
Sec |  |
SJYR01036895.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01001322.1 |  |
 |
3.199-20.789 (-) |  |
 |
 |
 |
 |
|
| SelO | ||||||||||||
| SelO |  |
Sec |  |
SJYR01002254.1 |  |
 |
325.440-344.530 (+) |  |
 |
 |
 |
 |
| SelP | ||||||||||||
| SelP |  |
Sec |  |
SJYR01002566.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01002566.1 |  |
 |
365.231-359.708 (-) |  |
 |
 |
 |
 |
|
| SelR | ||||||||||||
| SelR1 |  |
Sec |  |
SJYR01001528.1 |  |
 |
189.017-193.966 (-) |  |
 |
 |
 |
 |
| SelR2 |  |
Cys |  |
SJYR01000378.1 |  |
 |
103.402-117.923 (-) |  |
 |
 |
 |
 |
| SelR3 |  |
Cys |  |
SJYR01001010.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
- |  |
SJYR01000020.1 |  |
 |
699.131-765.343 (+) |  |
 |
 |
 |
 |
|
 |
- |  |
SJYR01001010.1 |  |
 |
24.614-106.809 (+) |  |
 |
 |
 |
 |
|
| SelS | ||||||||||||
| SelS |  |
Sec |  |
SJYR01003258.1 |  |
 |
291.560-299.601 (-) |  |
 |
 |
 |
 |
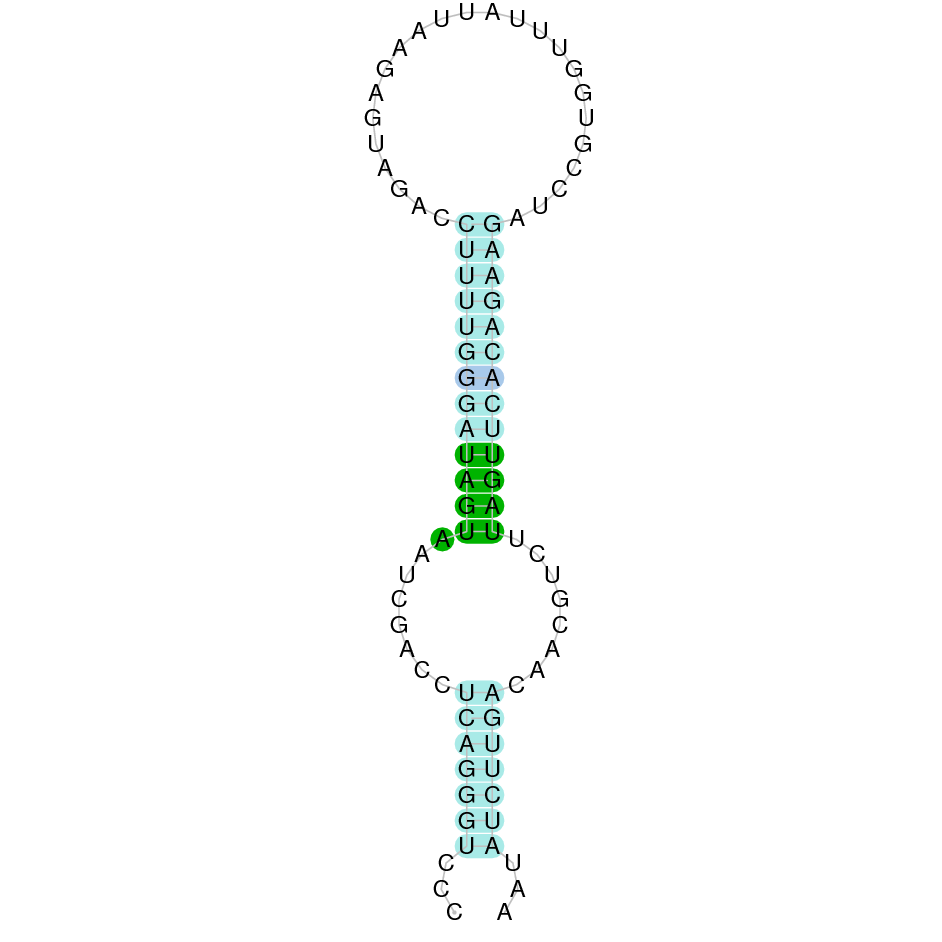
| SelU | ||||||||||||
| SelU1 |  |
Cys |  |
SJYR01001912.1 |  |
 |
362.613-366.415 (-) |  |
 |
 |
 |
 |
| SelU2 |  |
Cys |  |
SJYR01001714.1 |  |
 |
148.779-167.520 (+) |  |
 |
 |
 |
 |
| SelU3 |  |
Cys |  |
SJYR01001383.1 |  |
 |
271.344-273.745 (-) |  |
 |
 |
 |
 |
| TR | ||||||||||||
| TR1 |  |
Sec |  |
SJYR01000959.1 |  |
 |
701.737-739.796 (+) |  |
 |
 |
 |
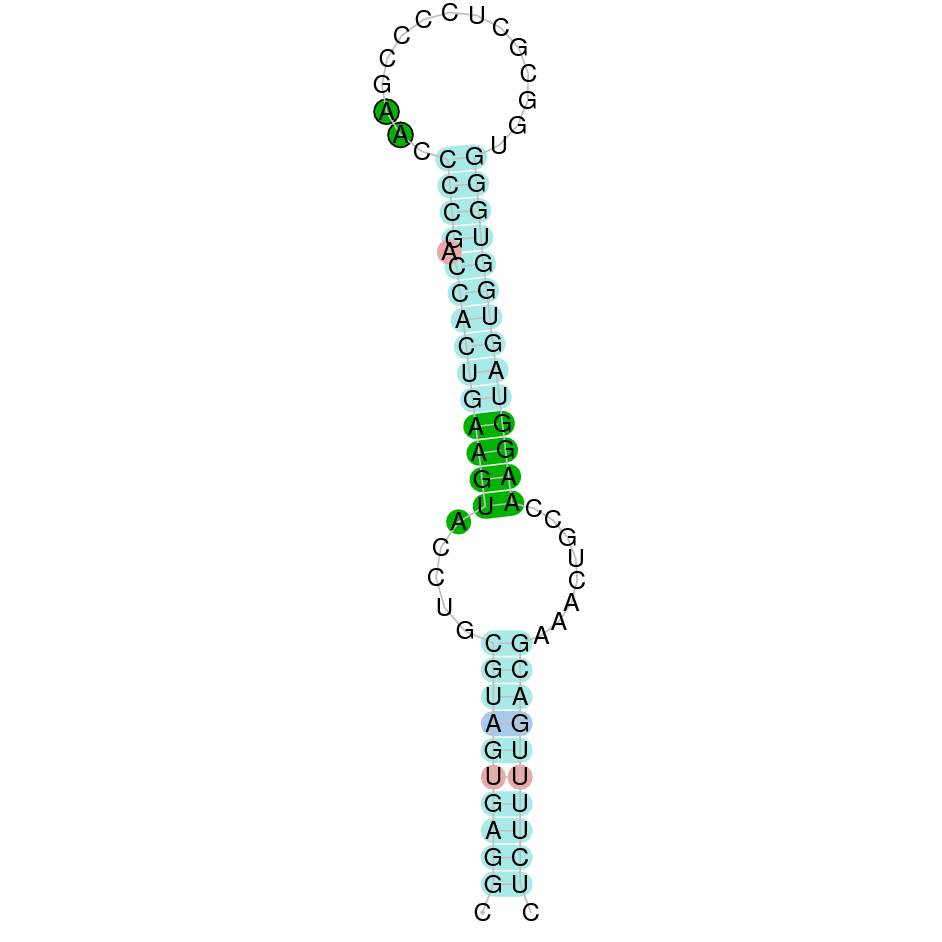 |
| TR2 |  |
Sec |  |
SJYR01003863.1 |  |
 |
- |  |
 |
 |
 |
 |
 |
Sec |  |
SJYR01003863.1 |  |
 |
696.86-94.531 (-) |  |
 |
 |
 |
 |
|
| TR3 |  |
Sec |  |
SJYR01001222.1 |  |
 |
509.806-565.183 (+) |  |
 |
 |
 |
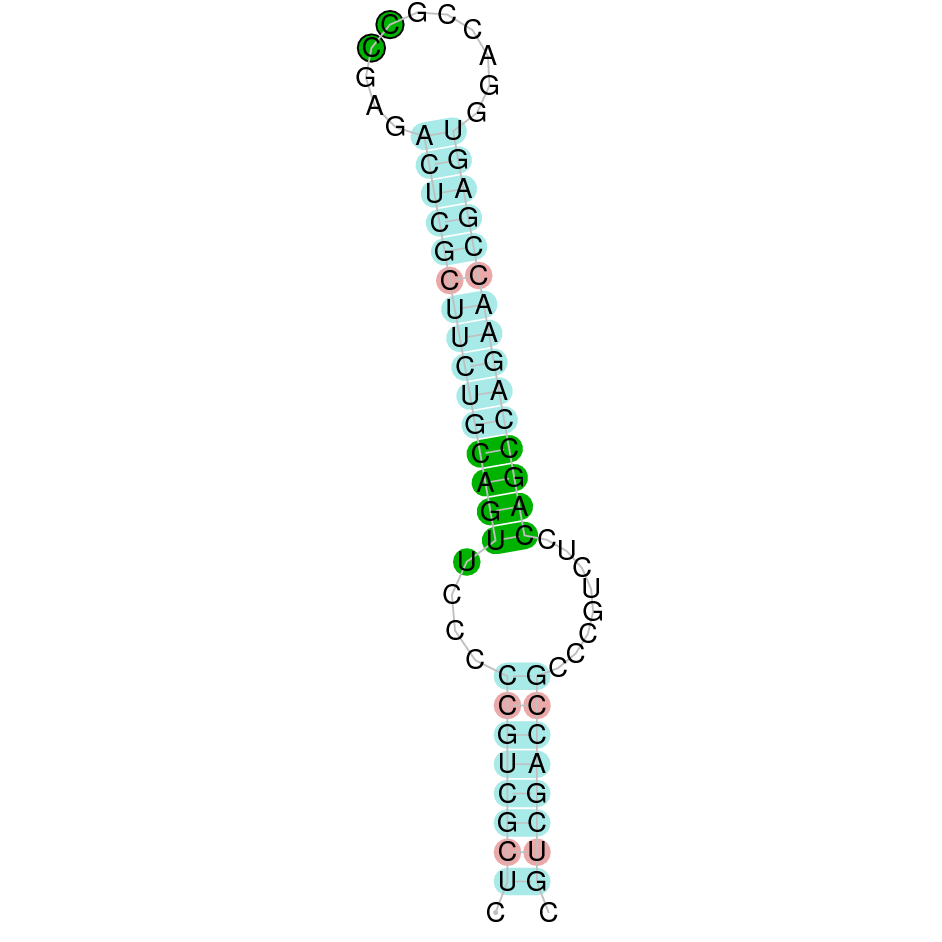 |
| Selenoprotein machinery | ||||||||||||
| eEFSec |  |
- |  |
SJYR01000075.1 |  |
 |
1.626.365-1.740.483 (-) |  |
 |
 |
 |
 |
| SBP2 |  |
Sec |  |
SJYR01003105.1 |  |
 |
262.147-287.310 (-) |  |
 |
 |
 |
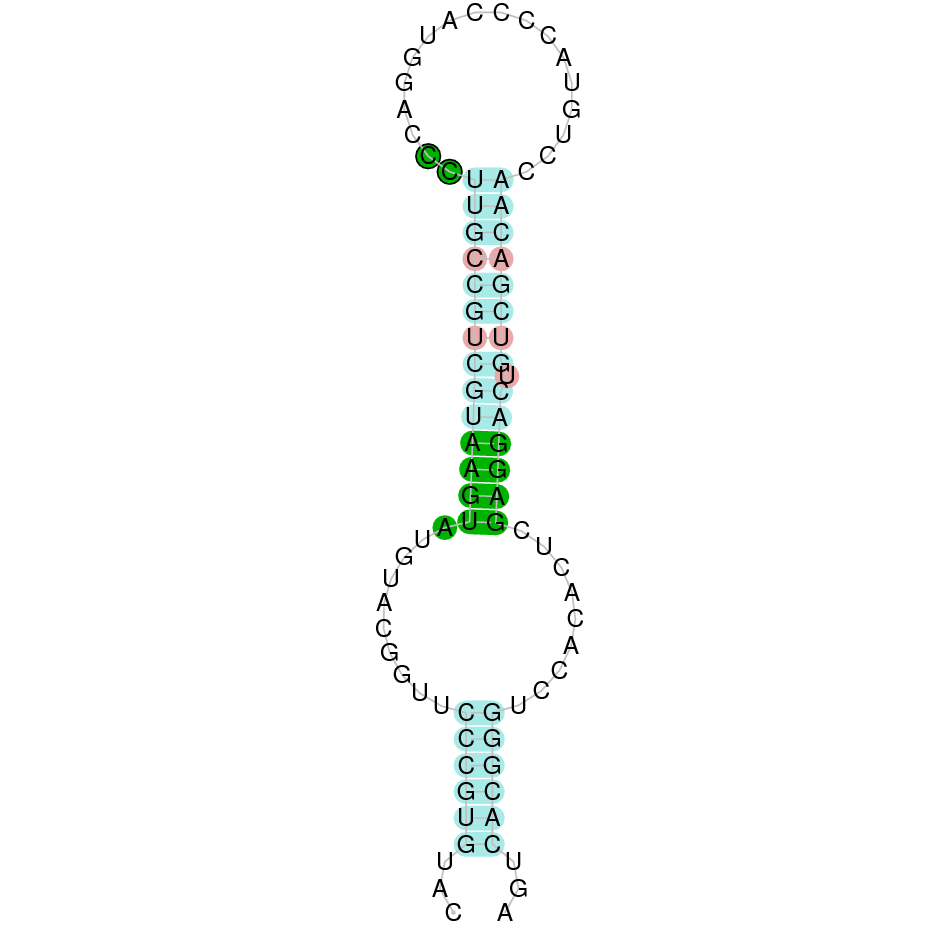 |
| SPS1 |  |
Tyr |  |
SJYR01001146.1 |  |
 |
26.372-26.266 (+) |  |
 |
 |
 |
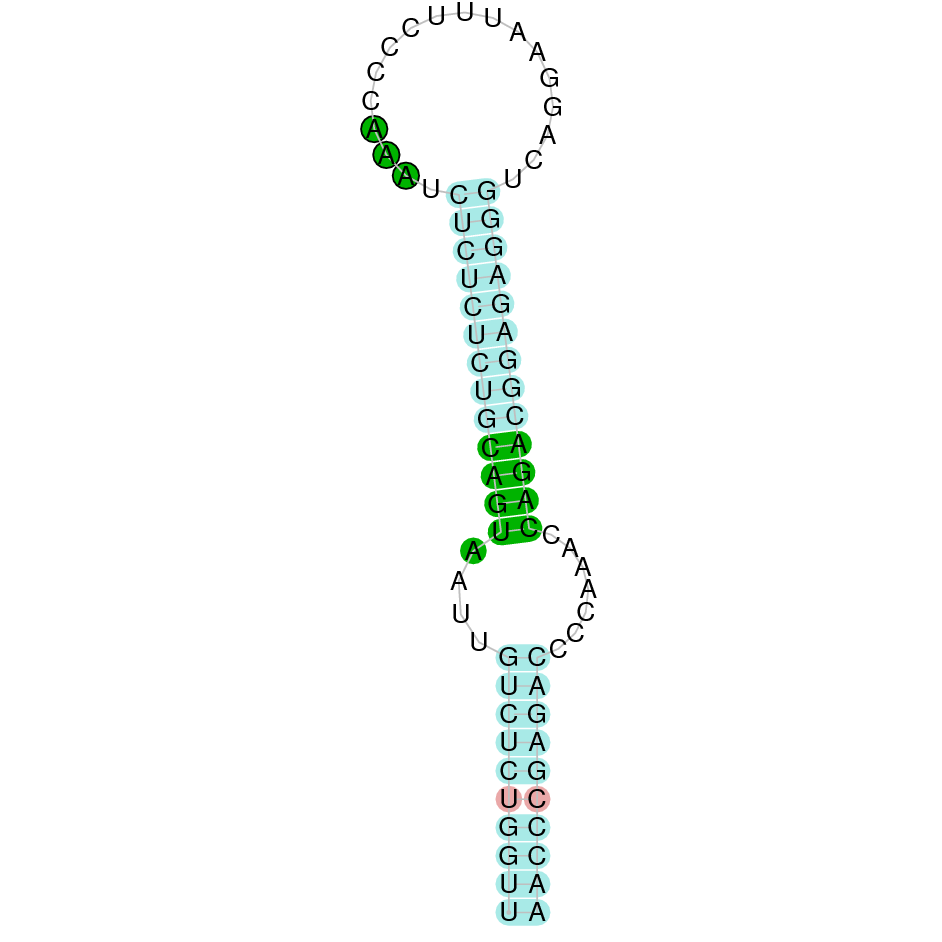 |
| SPS2 |  |
Sec |  |
SJYR01001146.1 |  |
 |
26.114-27.352 (+) |  |
 |
 |
 |
 |
Glutathione Peroxidase family (GPx)
In humans, the glutathione peroxidase family contains eight paralogs, but only five of them have selenocysteine [2]. All of them have the ability to catalyze the reduction of hydrogen peroxide by the use of glutathione as an electron donor. Its function in the human body also includes the detoxification of hydroperoxides and the maintaining cellular redox homeostasis [2]. In some of them, its activity is dependent on the selenocysteine [15], and they have a specific tissue expression. GPx1 was the first selenoprotein discovered in humans [16]. In addition, it turns out that GPX1 was the first mammalian protein discovered to have a Selenocysteine in the open reading frame. Until that moment it was thought that it coded for a stop codon instead of the amino acid selenocysteine. Last but not least, GPx6 homologs in some mammals have not conserved the Selenocysteine aminoacid and instead have a cysteine amino acid in the active site [2].
In the phylogenetic tree of Glutathione Peroxidase family, it can be seen that the predicted proteins GPx1 (predicted from H. sapiens), GPx2 (predicted from H. sapiens), GPx4 (predicted from H. sapiens), GPx7 (predicted from H. sapiens), GPx8 (predicted from H. sapiens) are classified together with their own query. This suggests that these proteins have been correctly predicted. However, the predicted proteins GPx3 (predicted from H. sapeins), GPx5 (predicted from B. taurus) and GPx6 (predicted from B. taurus) do not cluster together with their own queries. GPx1
GPx2
GPx3
GPx4
GPx5
GPx6
GPx7
GPx8
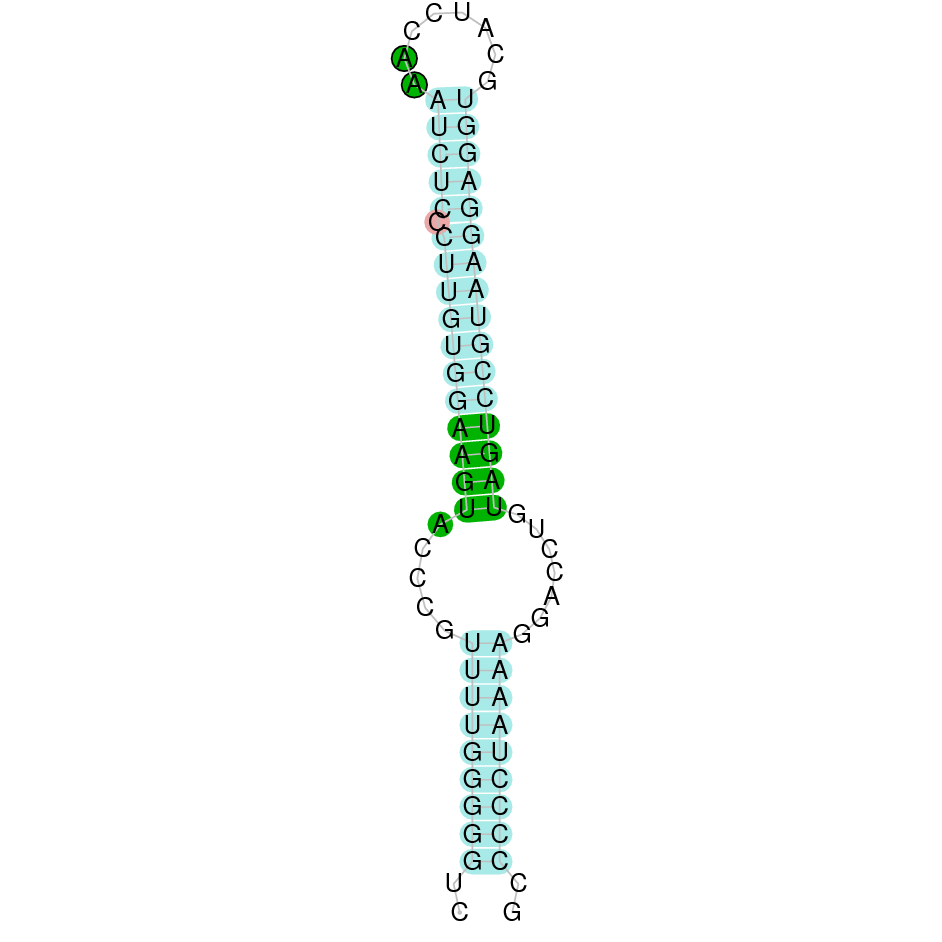
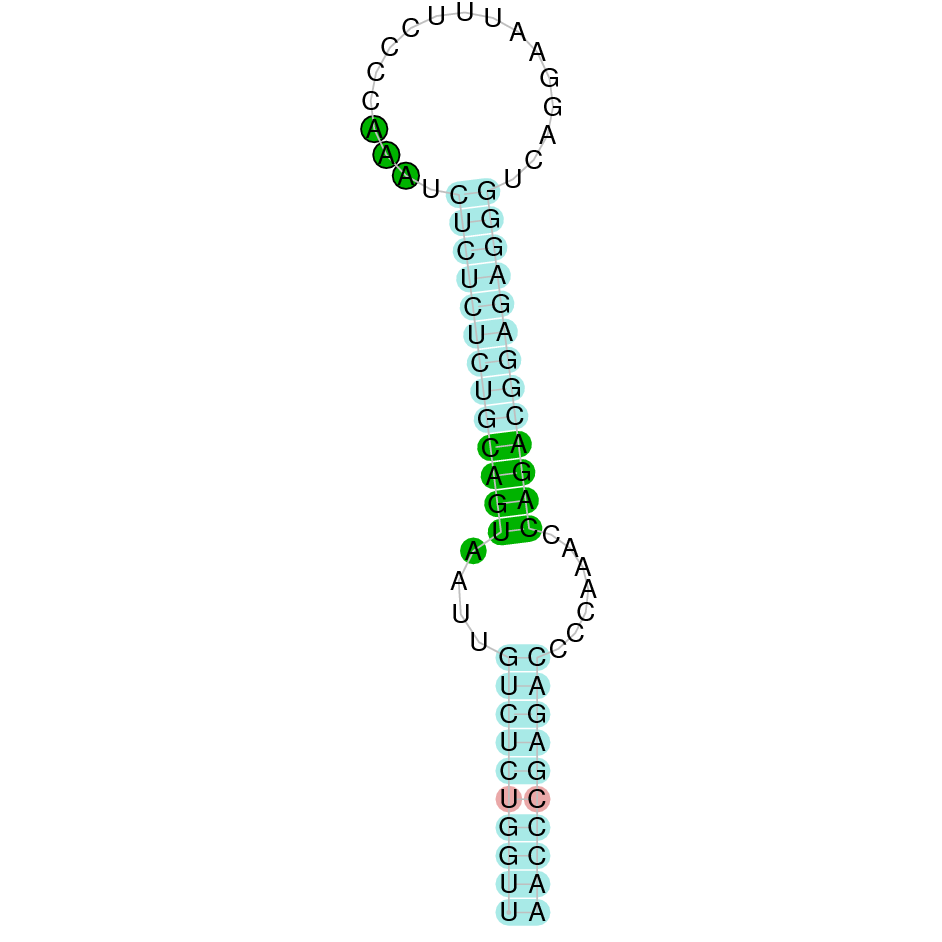
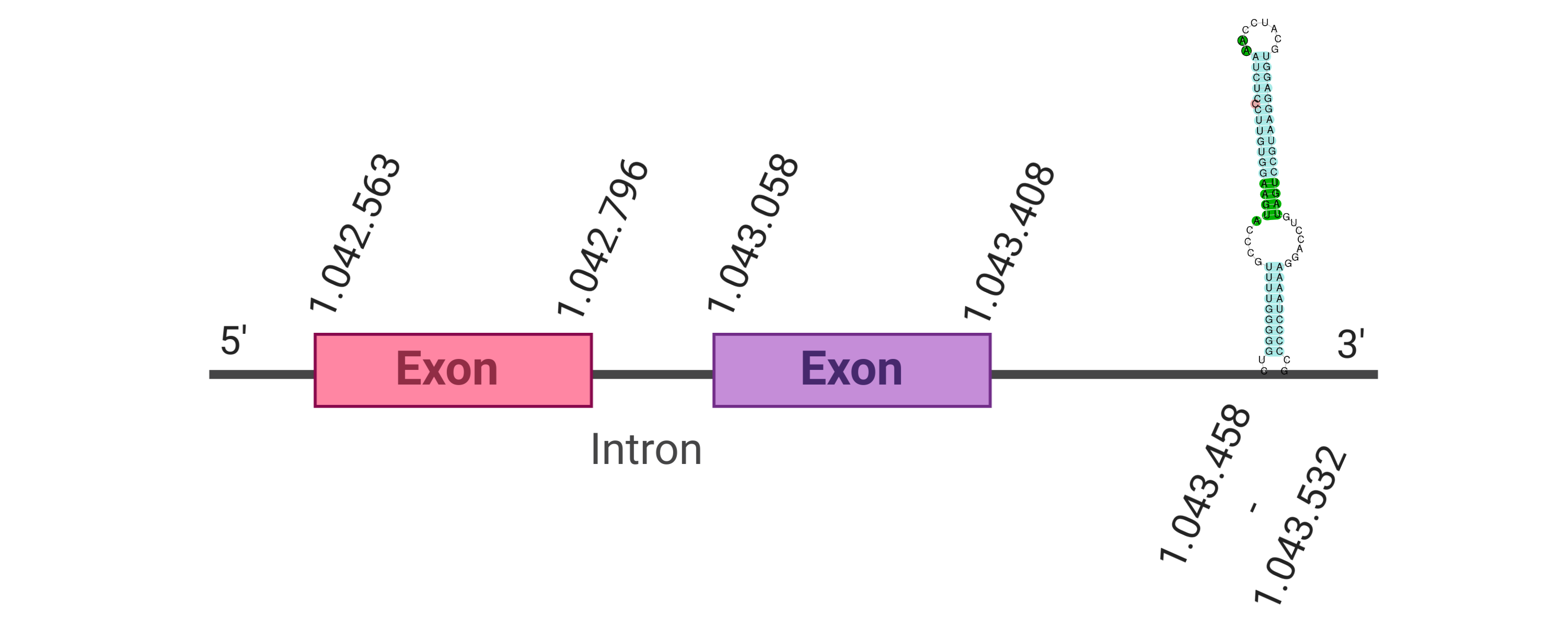
The protein GPx1 is located in the scaffold SJYR01000014.1 (it was chosen the hit with the lowest e-value: 1e-66). The percentage of identity between the query and the hit is 91%. The protein is located in the positive strand, between the nucleotide positions 1042563 and 1043408. In this protein, two exons were predicted. When running tcoffee, it was obtained a score of 998 and visualized quite a perfect alignment. Tcoffee aligns the human protein with the predicted protein, and it can be observed that both of them conserve the selenocysteine amino acid. Our predicted protein lacks the first methionine (in which it starts the protein), meaning that this region has not been correctly predicted. No protein was predicted using the Seblastian, but one secis element (grade A) was correctly predicted in the 3’ sequence of the protein.
Although the beginning of the protein could not be correctly predicted, the protein GPx1 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that GPx1 is a selenoprotein.
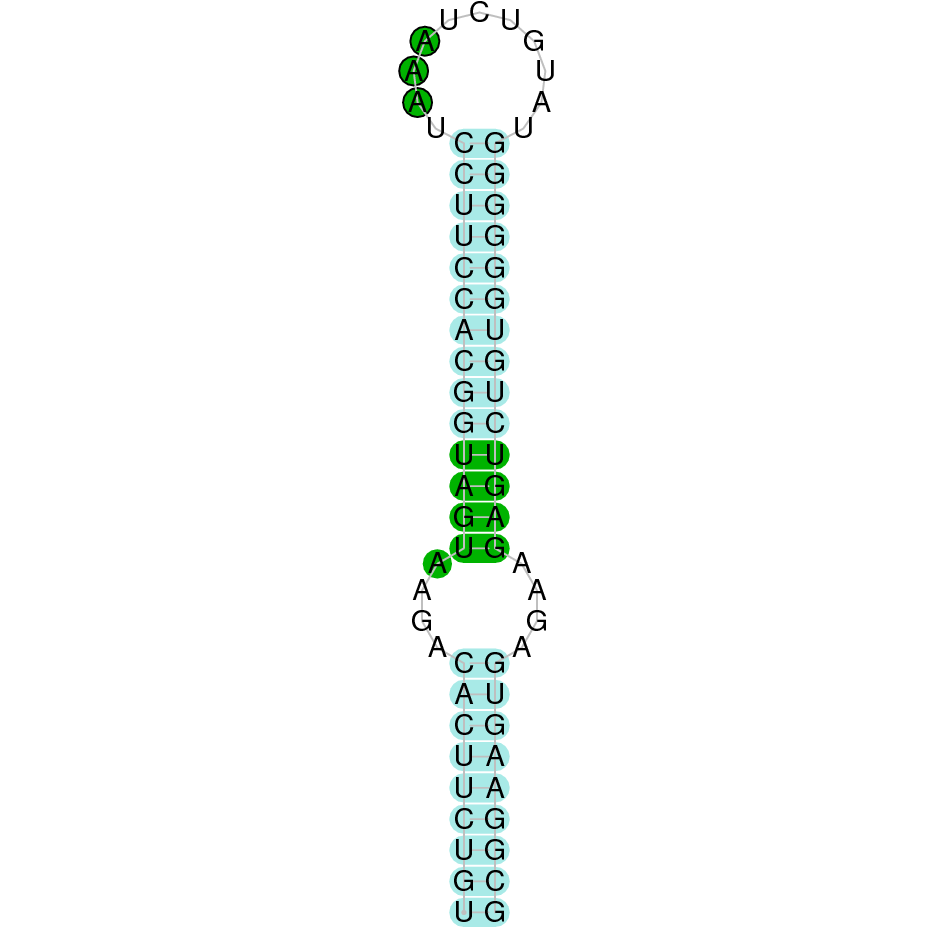
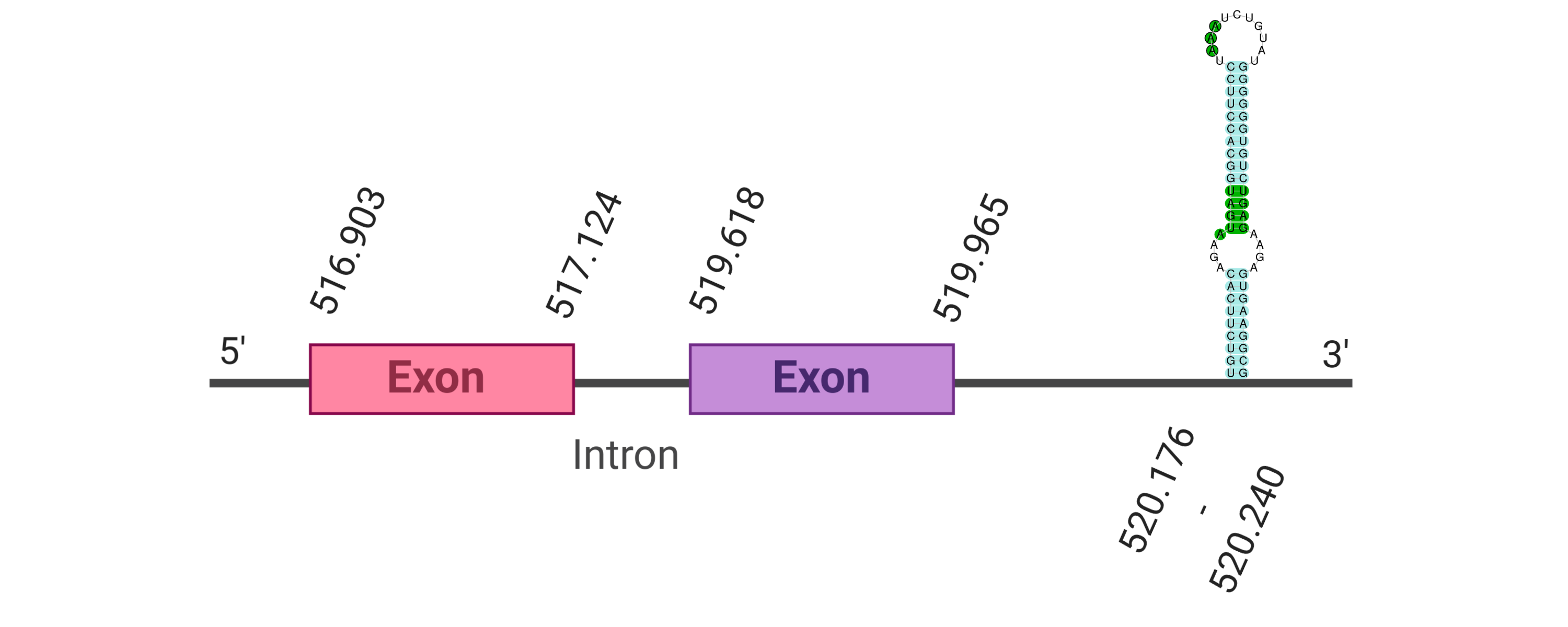
The protein GPx2 is located in the scaffold SJYR01000393.1 (it was chosen the hit with the lowest e-value: 7e-70). The percentage of identity between the query and the hit is 95%. The protein is located in the positive strand between the nucleotide positions 516903 and 519965. In this protein, two exons were predicted. In the tcoffee it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein, and it can be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been predicted correctly. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein GPx2 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that GPx2 is a selenoprotein.
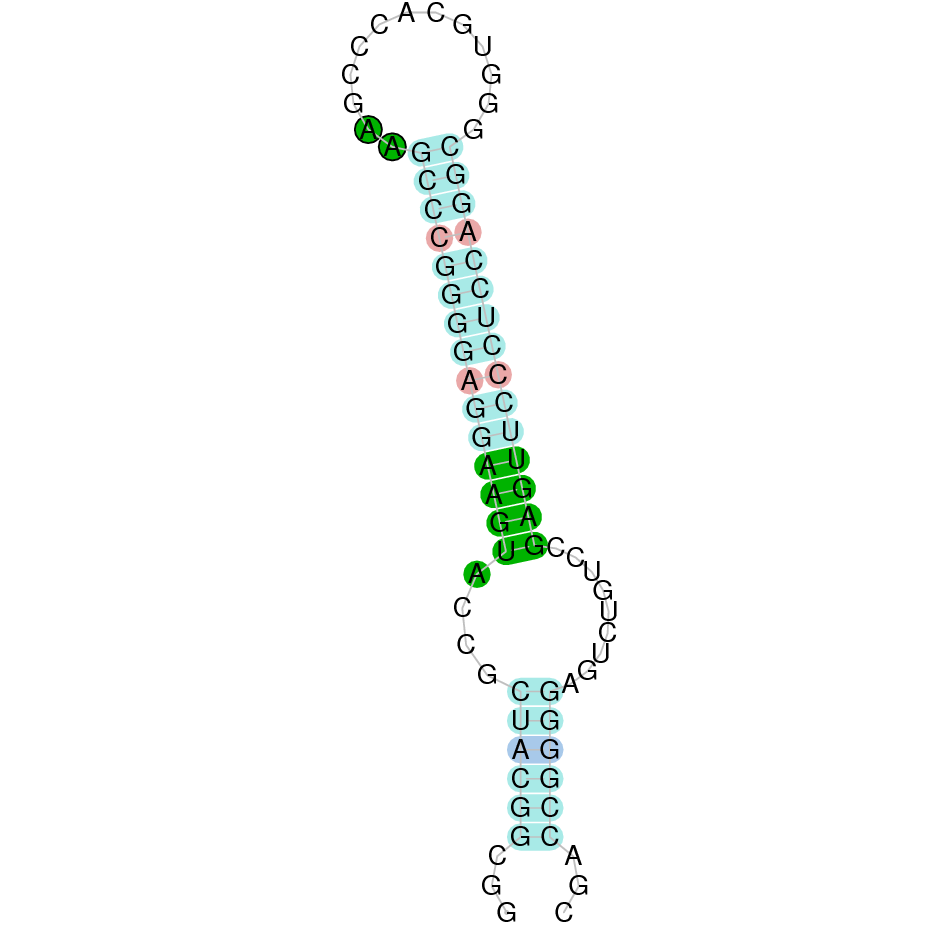
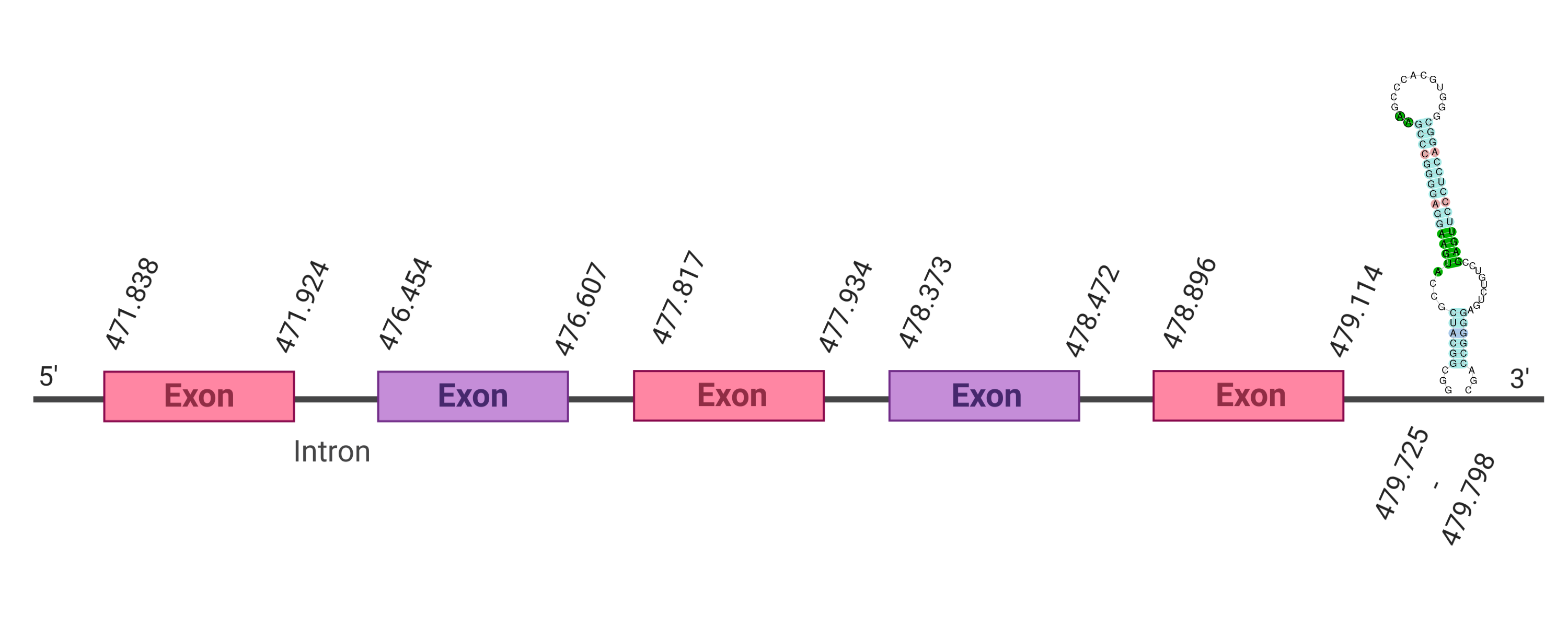
The protein GPx3 is located in the scaffold SJYR01000531.1 (the hit with the lowest e-value was chosen: 1.12e-35). The percentage of identity between the query and the hit is 79%. The protein is located in the positive strand between the nucleotide positions 471838 and 479114. In this protein, five exons were predicted. When running tcoffee, it was obtained a score of 1000, and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it can be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been predicted correctly. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein GPx3 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that GPx3 is a selenoprotein.
For this protein, different hits were evaluated. The first one, the one with the lowest e-value (5e-54), was located in the scaffold SJYR01000491.1. When running tcoffee, it was obtained the poor score of 787 and visualized a bad alignment with gaps inside the protein sequence. As the human protein GPx4 could not be correctly predicted in the genome of our animal, we decided to evaluate the second hit of tblastn.
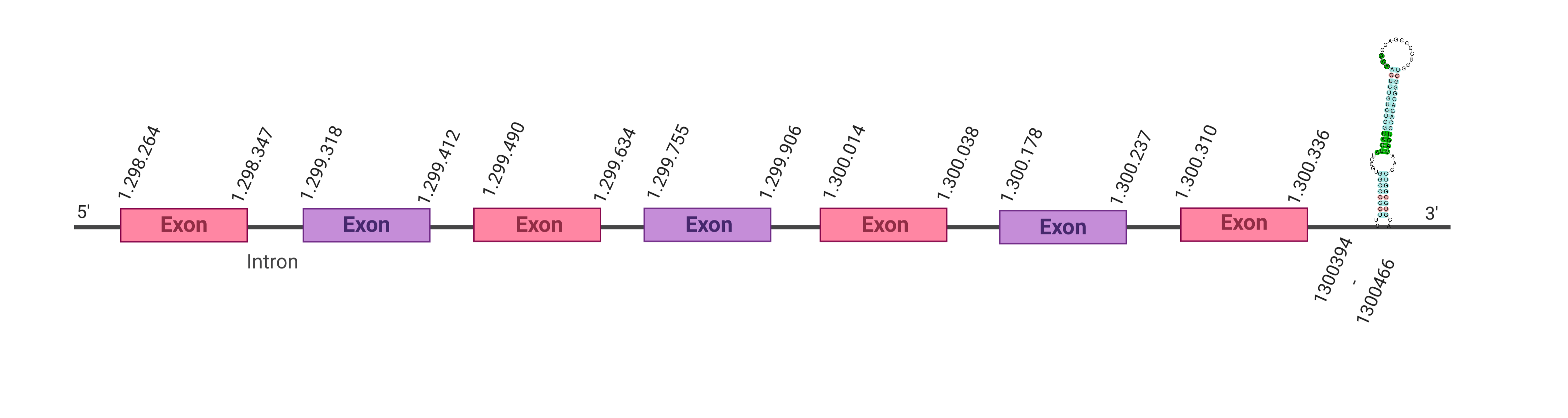
The second hit analyzed was located in the scaffold SJYR01000970.1, and had an e-value of 2.54e-49. The percentage of identity between the query and the hit is 65%. The protein is located in the positive strand, between the nucleotide positions 1298264 and 1300336. In this protein, seven exons were predicted. When running tcoffee, it was obtained a score of 1000, and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it can be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been predicted correctly. Using Seblastian, a selenoprotein from Bubalus bubalis was predicted. This protein has five exons, and it is located in the positive strand. One secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein GPx4 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that GPx4 is a selenoprotein.
For this protein different hits were evaluated. The first one was located in the scaffold SJYR01000174.1 (the hit with the lowest e-value was chosen: 1.71e-30). When running tcoffee, a score of 973 was obtained and visualized a bad alignment with gaps inside the protein sequence. As the GPx5 human protein was not correctly predicted in the genome of our animal, the second hit was evaluated.
The second hit analyzed was located in the scaffold SJYR01000014.1 and had an e-value of 2.19e-30. Despite having a promising e-value, the percentage of identity was only 49%. The predicted protein is located in the positive strand, between the nucleotide positions 99577 and 100389. In this protein, two exons were predicted. When running tcoffee, a score of 990 was obtained. Although it is not the best alignment, it is better than the previous one. The alignment obtained still had some gaps inside the protein sequence and the predicted protein lacks the methionine amino acid at the beginning, meaning that this region has been lost. The human protein GPx5 did not contain Selenocysteine (contains cysteine) while the predicted GPx5 protein in K. ellipsiprymnus did.
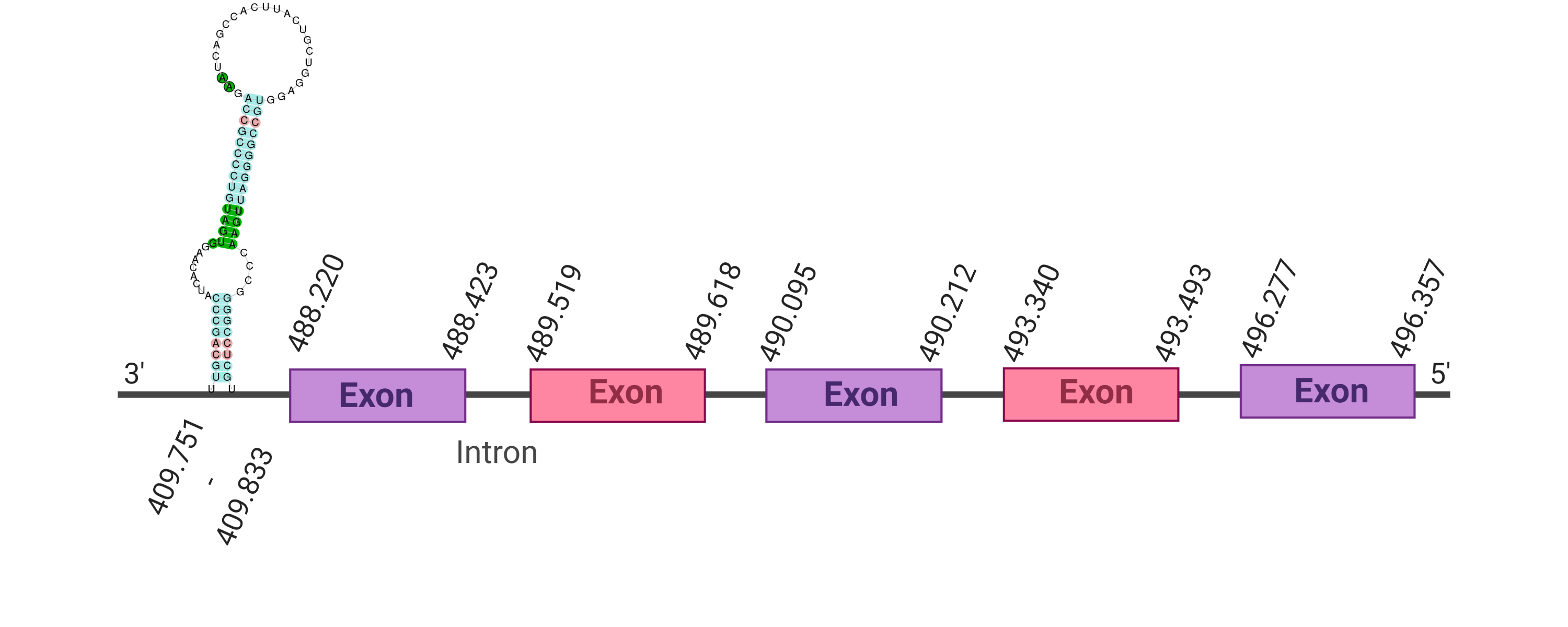
As the predicted protein from the human protein GPx5 was not as good as it would be expected, the GPx5 protein in K. ellipsiprymnus was predicted again from the GPx5 Bos taurus’ protein (Cow). By doing so, the protein GPx5 is located in the scaffold SJYR01000174.1 (the hit with the lowest e-value obtained: 1.83e-37). The percentage of identity between the query and the hit is 89%. The protein is located in the negative strand between the nucleotide positions 488423 and 516694. In this protein, five exons were predicted. When running tcoffee, a score of 999 was obtained, and visualized quite a perfect alignment. However, the predicted protein of K. ellipsiprymnus seems to be larger than the protein sequence of the cow (the cow selenoprotein aligns in the middle of the predicted protein in K. ellipsiprymnus). The cow selenoprotein GPx5 does not contain selenocysteine. No protein was predicted using the Seblastian, but one secis element (grade B) in the negative strand was correctly predicted in the 3’ sequence of the protein.
The protein GPx5 in K. ellipsiprymnus does not contain a selenocysteine but it still contains a Secis element. This suggest that nowadays K. ellipsiprymnus’ GPx5 is not a selenoprotein.
For this protein, different hits were evaluated. The first one, the one with the lowest e-value (2e-35), was located in the scaffold SJYR01000174.1. When running tcoffee, it was obtained a score of 969 and visualized a bad alignment with gaps inside the protein sequence. Also, it was not correctly predicted the upper region of the protein. As the human GPx6 protein was not correctly predicted in K. ellipsiprymnus, the second hit was evaluated.
The second hit analyzed was located as well in the scaffold SJYR01000174.1, and had an e-value of 5.22e-27. When running tcoffee, it was obtained the same score (969) again and visualized a bad alignment with gaps inside the protein sequence (probably because the same protein was predicted). The human protein GPx6 did not contain selenocysteine (it contains a cysteine) while the predicted GPx6 protein in K. ellipsiprymnus did.
As the predicted protein from the human protein GPx6 was not as good as it would be expected, the GPx6 protein in K. ellipsiprymnus was predicted again from the GPx6 Bos taurus’ protein (Cow). By doing so, the protein GPx6 is located in the scaffold SJYR01000174.1 (the hit with the lowest e-value was chosen: 1.21e-39). In the tcoffee, a sore of 949 was obtained and visualized a bad alignment with gaps inside the protein sequence. Therefore, the second hit of tblastn evaluating the GPx6 Bos taurus’ protein was used.
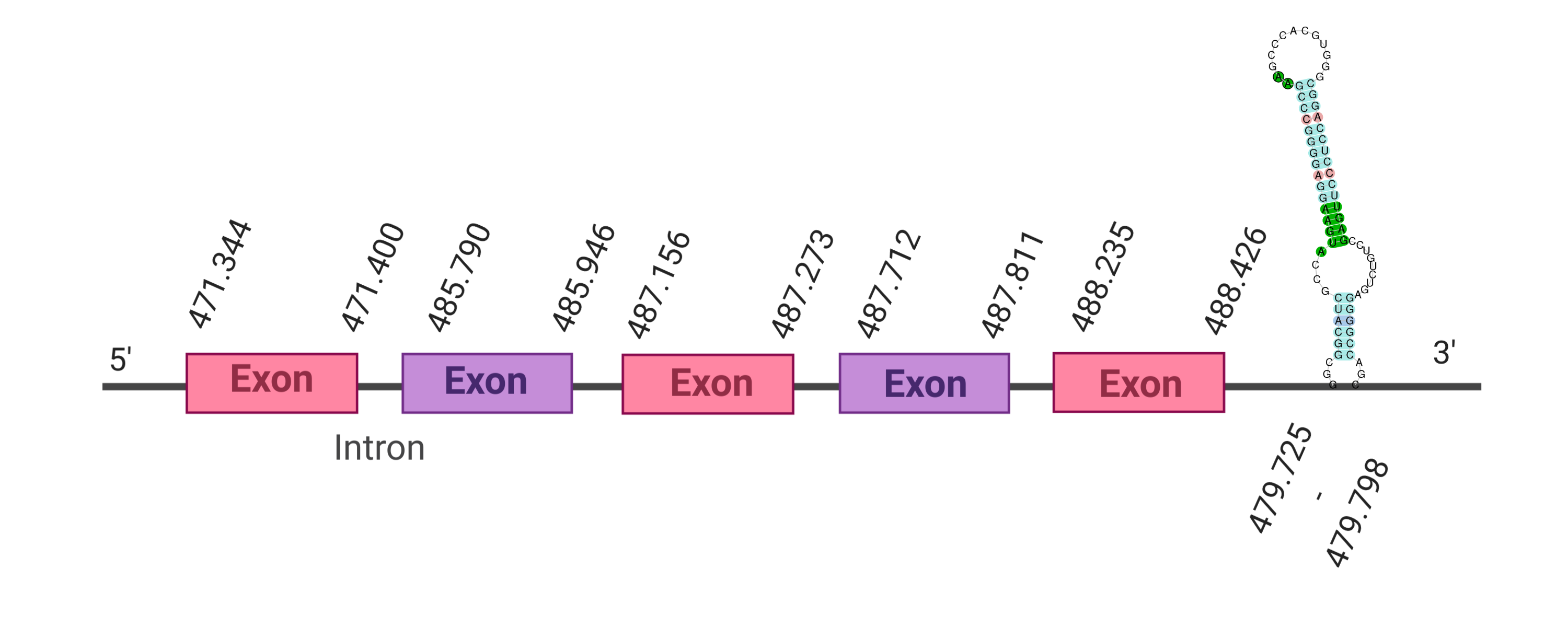
The second hit was located as well in the scaffold SJYR01000531.1 and had an e-value of 1.04e-28. The percentage of identity between the query and the hit is 69%. The predicted protein is located in the positive strand between the nucleotide positions 471344 and 488426. In this protein, five exons were predicted. When running tcoffee, it was obtained a score of 999 and visualized quite a perfect alignment. The cow selenoprotein GPx6 does contain selenocysteine, while the predicted GPx6 protein lacks the methionine amino acid at the beginning, meaning that the beginning of the protein sequence has been lost. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein GPx6 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that GPx6 is a selenoprotein.
For this protein, different hits were evaluated. The first one was located in the scaffold SJYR01000129.1 (it was chosen the hit with the lowest e-value: 7.60e-57). When running tcoffee, it was obtained a poor score of 919 and visualized a bad alignment with gaps inside the protein sequence. As the protein GPx7 could not be correctly predicted from the human GPx7 protein, the second hit was evaluated.
The second hit was located in the scaffold SJYR01000065.1, and had an e-value of 8.70e-53. The percentage of identity between the query and the hit was 88%. The predicted protein is located in the positive strand, between the nucleotide positions 516812 and 525517. In this protein, three exons were predicted. When running tcoffee, it was obtained a score of 998 and visualized quite a perfect alignment. Tcoffee aligns the human protein with the predicted protein and, it can be observed that both of them contain a cysteine amino acid. The predicted GPx7 protein lacks the methionine amino acid at the beginning, meaning that the starting point of this protein has been lost. No protein was predicted using the Seblastian. Additionally, any secis element was found in the positive strand.
Taken all together, the protein GPx7 in K. ellipsiprymnus does not contain a selenocysteine amino acid nor a Secis element. All these signs would suggest that nowadays GPx7 is a cysteine homologous protein.
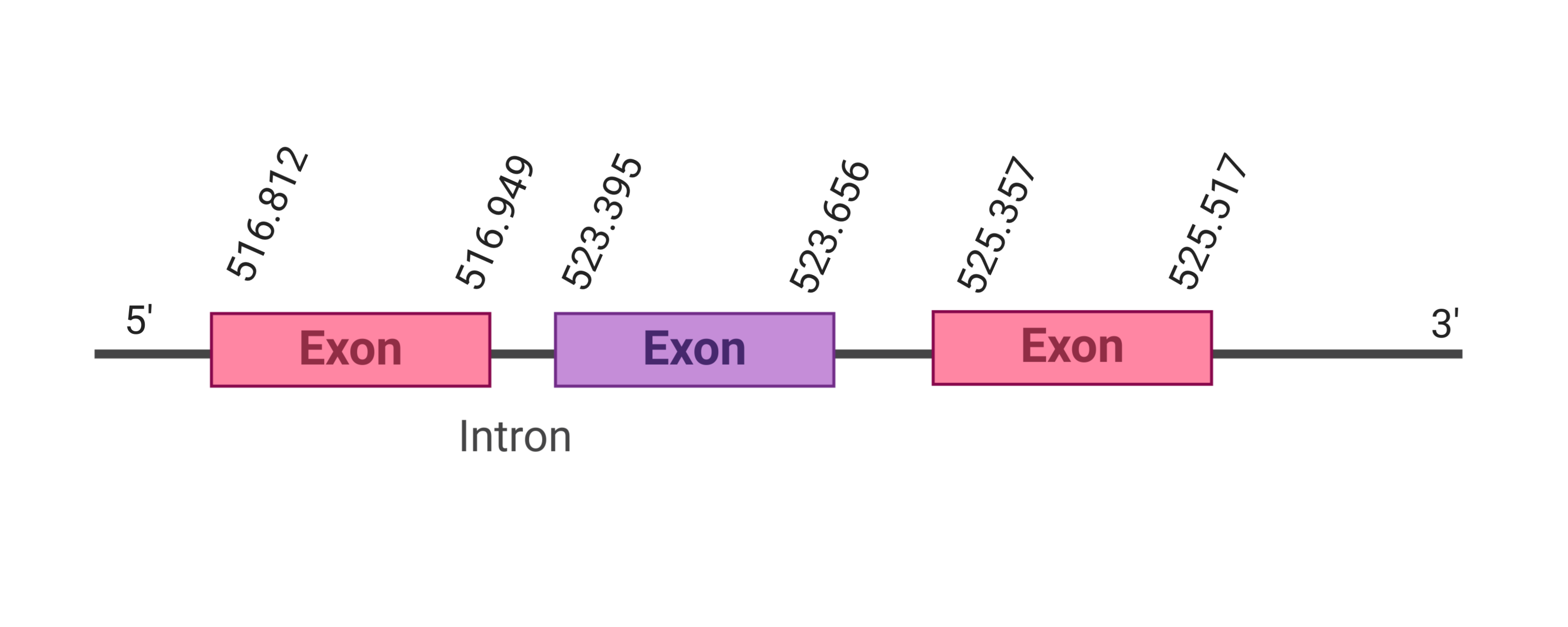
The protein GPx8 is located in the scaffold SJYR01000057.1 (it was chosen the hit with the lowest e-value: 1.33e-45). The percentage of identity between the query and the hit is 90%. The protein is located in the negative strand between the nucleotide positions 1105599 and 1101599. In this protein, three exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and, it could be observed that both of them have a cysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been predicted correctly. No protein was predicted using the Seblastian. Additionally, any secis element was found in the negative strand.
Taken all together, the protein GPx8 in K. ellipsiprymnus does not contain a selenocysteine amino acid nor a Secis element. All these signs would suggest that nowadays GPx8 is a cysteine homologous protein.
Thyroid Hormone Deodinases (DI)
The Thyroid hormone deiodinases family contains three paralogous proteins (DI1, DI2 and DI3). Like GPx family, these proteins have a different tissue localization and expression [2]. All of them modulate the activation or inactivation of the thyroid hormone. Both DI1 and DI3 catalyze the removal of iodine of T4. Therefore, they produce T3 in plasma or peripheral tissues, respectively. On the other hand, DI2 catalyzes this reaction in specific tissues activating the thyroid hormone[2].
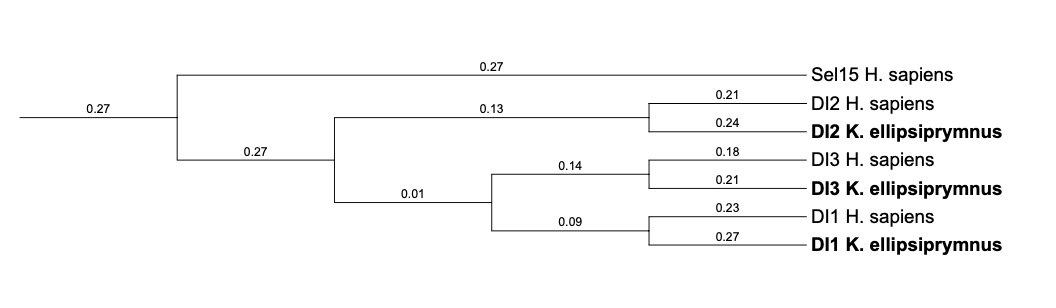
In the phylogenetic tree of Thyroid Hormone Deodinases family, it can be seen that the selenoproteins DI1, DI2 and DI3 predicted in K. ellipsiprymnus are classified together with their own queries. It suggests that the proteins have been predicted correctly.
DI1
DI2
DI3
MsrA: methionine-S-sulfoxide reductase
MsrA (Methionine sulfoxide reductase A) is responsible for the reduction of the enantiomer methionine-S-sulfoxide [2] giving place to methionine. It can reduce this amino acid when it is present in the free form or proteins. Depending on the alternative splicing that takes place, it can be located in the mitochondria, in the nucleus or the cytosol[17]. Msr proteins have an essential function in protecting the cell from oxidative stress.
Sel15
Selenoproteins 15 (Sel15) are thiol-disulfide oxidoreductases located in the endoplasmic reticulum (ER) which are implicated in protein folding [2]. These proteins are present in many mammalian tissues, but their highest level of expression is in the prostate, liver, kidney and testis. Sel15 forms a complex with UGGT, a chaperone located in the ER that contributes to the regulation of the calnexin pathway. This protein quality control pathway participates in the folding of N-linked glycoproteins in the ER. Therefore, Sel15 is involved in glycoproteins folding. Sel15 has also been thought to regulate redox homeostasis in the ER and to mediate the cancer prevention effect of Se [2].
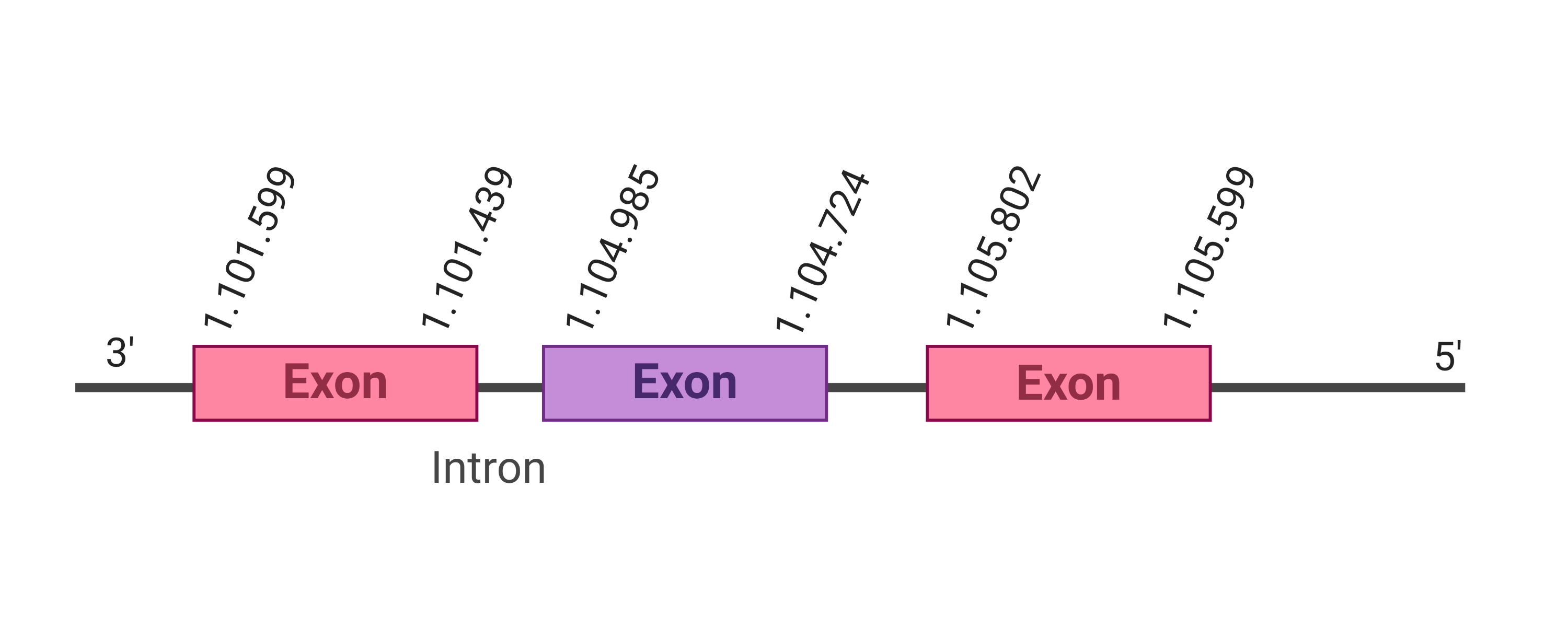
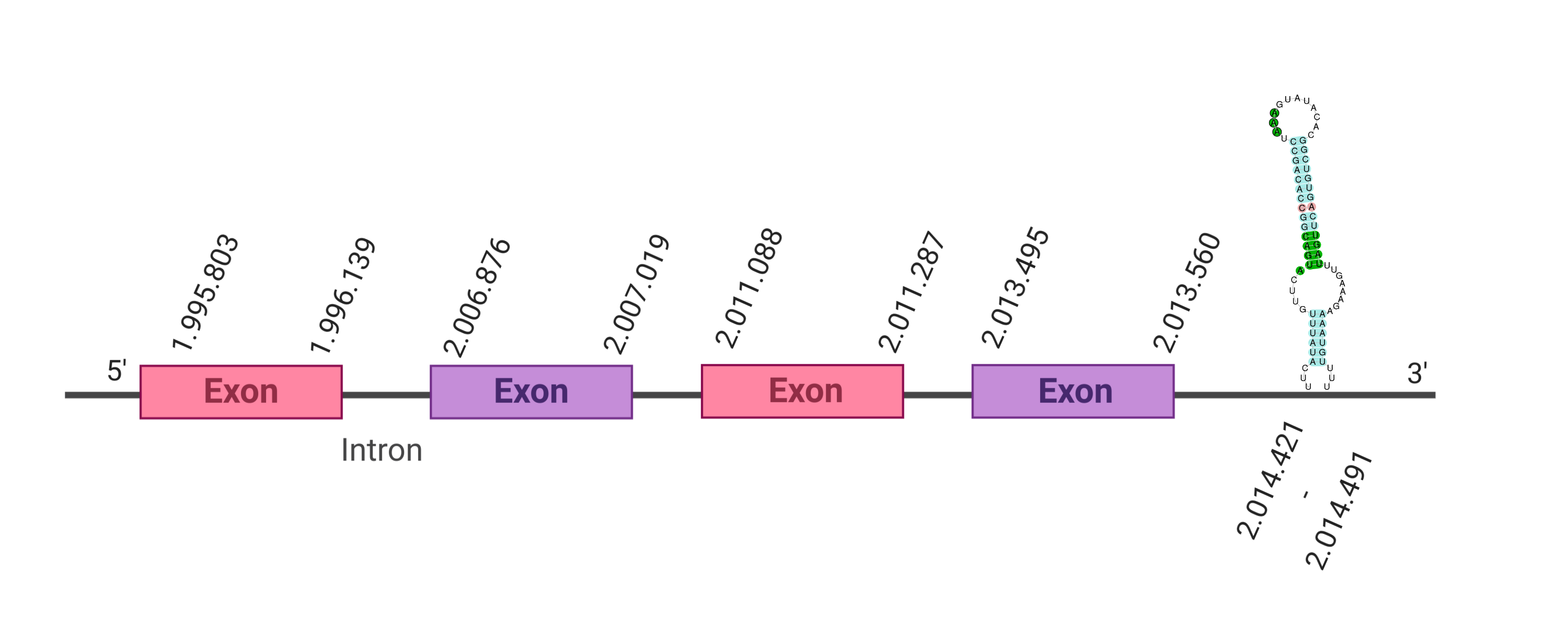
The protein DI1 is located in the scaffold SJYR01000065.1 (it was chosen the hit with the lowest e-value: 7.87e-48). The percentage of identity between the query and the hit is 83%. The protein is located in the positive strand between the nucleotide positions 1995803 and 2013560. In this protein, four exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and, it could be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. Using Seblastian, a selenoprotein from Ovis aries was predicted. This protein has four exons, and it is located in the positive strand. One secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein DI1 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that DI1 is a selenoprotein.
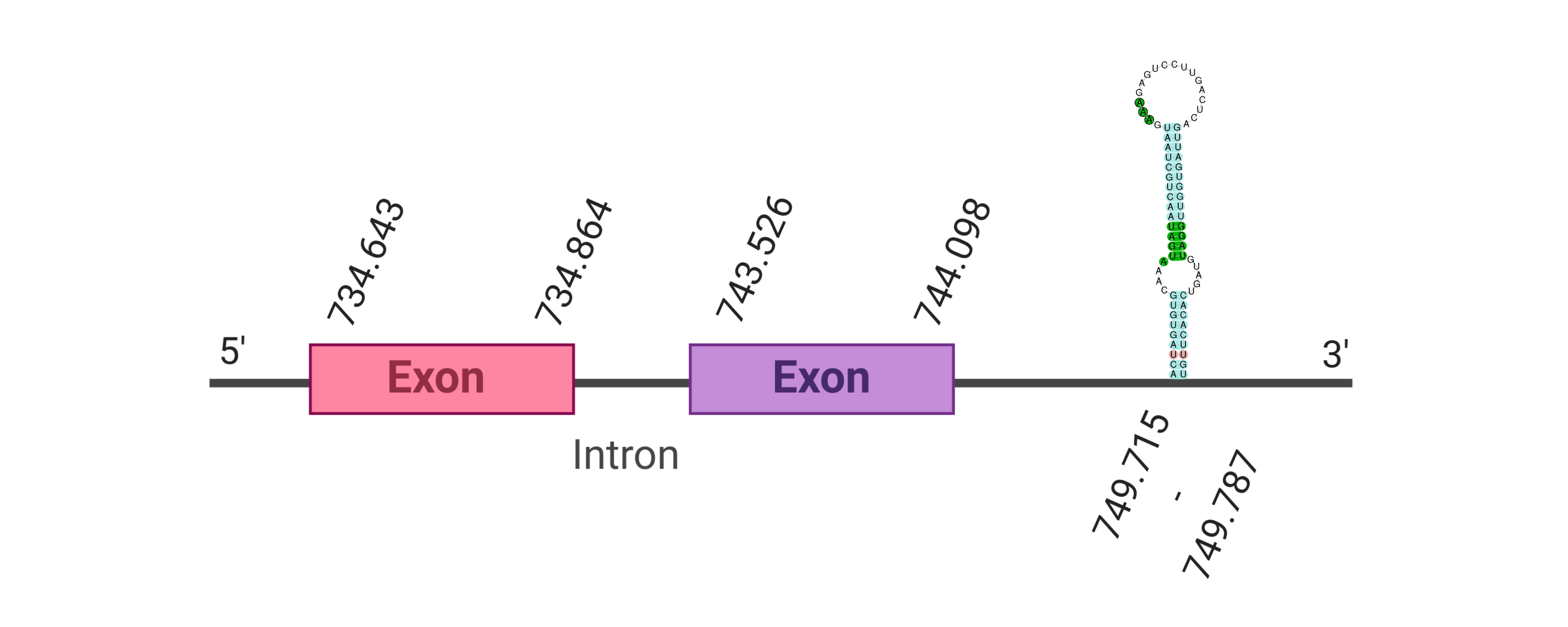
The protein DI2 is located in the scaffold SJYR01000770.1 (it was chosen the hit with the lowest e-value: 2.78e-112). The percentage of identity between the query and the hit is 89%. The protein is located in the positive strand between the nucleotide positions 734643 and 744098. In this protein, two exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and, it could be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein DI2 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that DI2 is a selenoprotein.
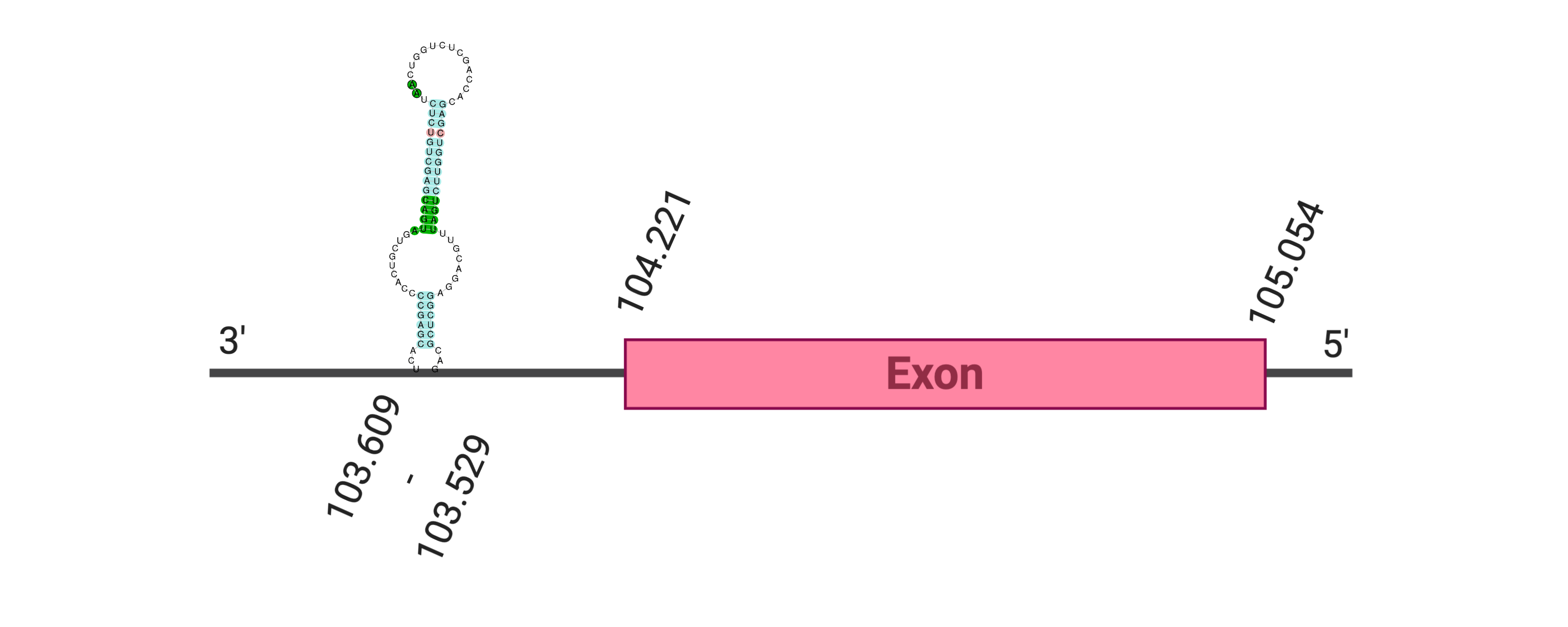
The protein DI3 is located in the scaffold SJYR01002697.1 (it was chosen the hit with the lowest e-value: 1.76e-168). The percentage of identity between the query and the hit is 94%. The protein is located in the reverse strand between the nucleotide positions 103529 and 104221. In this protein, one exon was predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and, it could be observed that both of them conserve the selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. Using Seblastian, a selenoprotein from Ovis aries was predicted. This protein has one exon, and it is located in the negative strand. One secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
The protein DI3 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that DI3 is a selenoprotein.
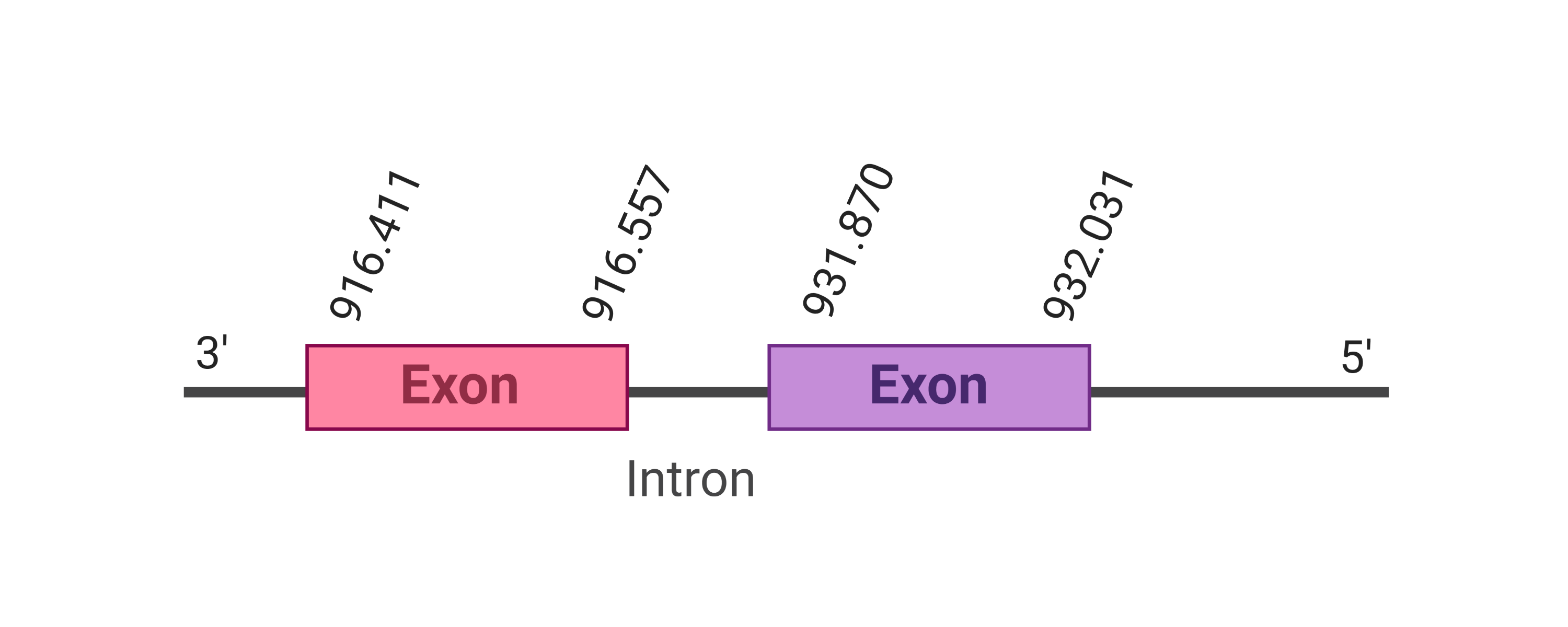
For this protein, different hits were evaluated. The first one, the one with the lowest e-value (3.27e-26), was located in the scaffold SJYR01026337.1. The predicted protein is located in the reverse strand between the nucleotide positions 1530 and 1691, and only one exon was predicted. When running tcoffee, it was obtained a score of 998. Despite the good score obtained, only the final sequence of the protein was predicted correctly (the first part is not predicted, including the first methionine).
In order to obtain a better prediction of the protein, the second hit was evaluated. The predicted protein was located in the scaffold SJYR01000661.1, in the negative strand, between the nucleotide positions 916411 and 932031. In this protein, two exons were predicted. When running tcoffee, it was obtained the same score (998). Like in the other hit, only the final part of the selenoprotein was correctly predicted. The human protein contains a cysteine, while both the predicted proteins (for the two hits) contain selenocysteine.
As the protein was not correctly predicted using the human selenoprotein as a query, it was predicted again using the MsrA Bos taurus’s protein (Cow). The first two hits obtained (the ones with the higher e-value) when using the cow selenoprotein are the same ones obtained with the human MsrA protein. When running tcoffee, it was obtained the same score.
To sum up, the same predicted protein was obtained when running tcoffee for two different hits placed in two different scaffolds. The same scenario happened when using MsrA Bos taurus’s protein in order to do the prediction. These results suggest that there has been a partial duplication (only the final sequence of the protein) of MsrA in K. ellipsiprymnus. No protein was predicted using the Seblastian. Additionally, any secis element was found in the negative strand.
Taken all together, the protein MsrA in K. ellipsiprymnus does not contain a selenocysteine amino acid nor a Secis element. All these signs would suggest that nowadays MsrA is not a selenoprotein.
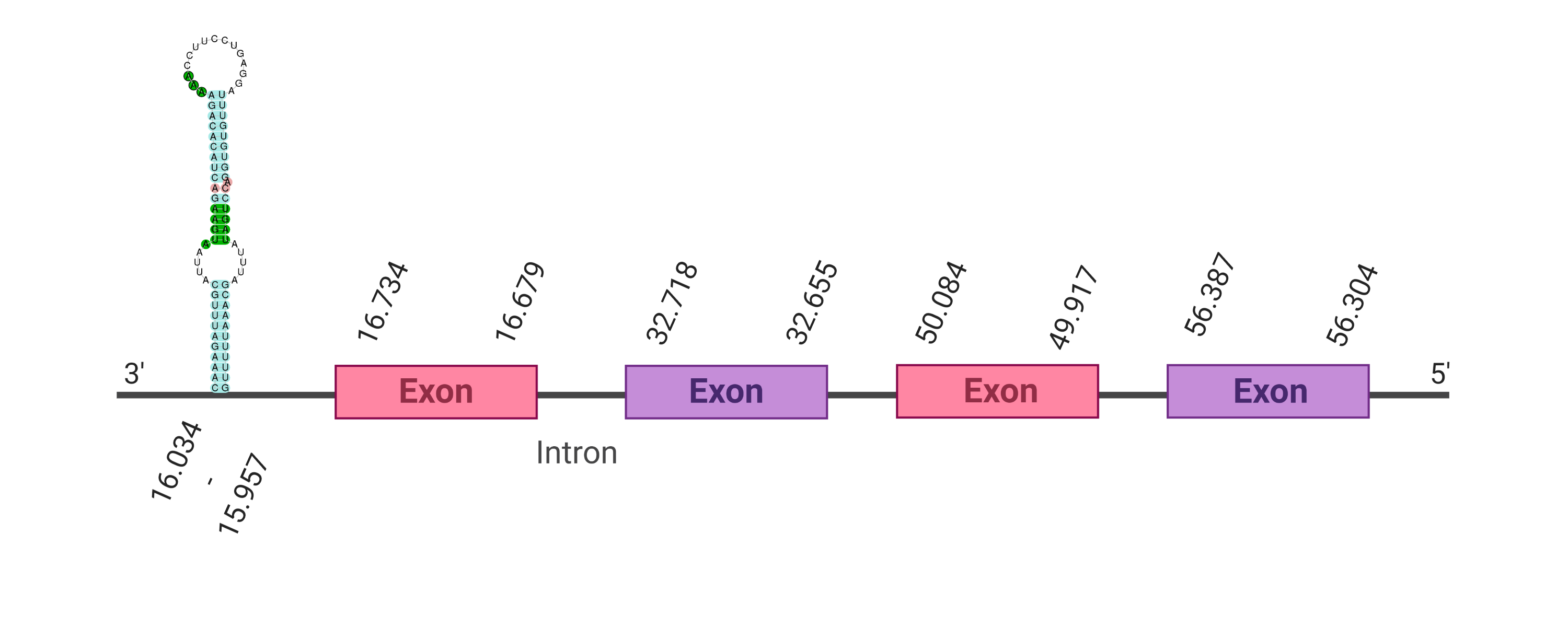
The protein Sel15 is located in the scaffold SJYR01001197.1 (it was chosen the hit with the lowest e-value: 6.43e-19). The percentage of identity between the query and the hit is 87%. The protein is located in the negative strand between the nucleotide positions 16679 and 56387. In this protein, four exons were predicted. When running tcoffee, it was obtained a score of 1000 and, visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that both the human protein Sel15 and the predicted protein in K. ellipsiprymnus conserve the selenocysteine. The predicted protein lacks the first methionine amino acid (it has a valine instead), meaning that the beginning of the protein sequence has not been correctly predicted. Using Seblastian, the selenoprotein Sel15 from Octodon degus was predicted. This protein has three exon, and it is located in the negative strand. One secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
The protein Sel15 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that Sel15 is a selenoprotein.
Thioredoxin-like family
Selenoprotein W (SelW), T (SelT), H (SelH), and V (SelV) belong to the redox (Rdx) family of selenoproteins: Thioredoxin-like Family. The members of this protein family possess a thioredoxin-like fold and are characterized by the presence of a conserved Cysx-x-Sec motif [18].
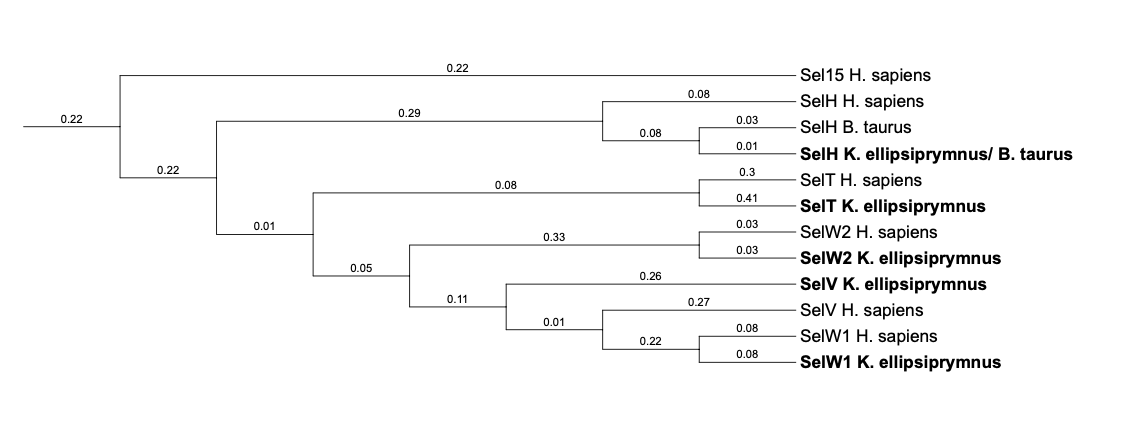
In the phylogenetic tree of Thioredoxin-like family, it can be seen that the predicted proteins SelT (predicted from H. sapiens), SelH (predicted from B. taurus), SelW2 and SelW1 (predicted prom H. sapiens) are classified together with their own query. This suggests that these proteins have been correctly predicted. However, the predicted proteins SelH (predicted from H. sapiens) and SelV (predicted from H. sapiens) do not cluster together with their own queries, meaning that these proteins are better predicted from B. taurus than H. sapens.
SelH
SelT
SelV
SelH is an oxidoreductase located in the nucleoli. Different analysis in mouse indicates that it has a high expression during the embryonic development, whereas it is low in adult tissues. SelH has a thioredoxin-like fold motif, a DNA-binding domain and it also has glutathione peroxidase activity. In addition, it plays a key role in the regulation of transcription of genes involved in de novo glutathione synthesis [2,19]. It may also have a regulatory role in redox homeostasis. SelH is thought to transduce oxidant signals by modulating gene expression [20].
Four hits were obtained when running tblastn, all of them located in the scaffold SJYR01002415.1. All hits were located in the positive strand. The first two hits had the same e-value (3.08e-15) and the same percentage of identity, and the last two hits also had the same e-value between them (1.42e-10) and the same percentage of identity.
The protein was predicted using both the first hit, which had an e-value of 3.08e-15 and the second hit, which had an e-value of 1.42e-10. In both of them we predicted the same number of exons (six exons) and the same positions for them. The tcoffee outcome obtained in both predicted proteins had a score of 733. Both the starting and the final region of the protein (including the first methionine) is correctly predicted. However, many gaps are present in between, maybe due to the fact that the K. ellipsiprymnus SelH protein have less exons than the human SelH protein. It can be observed that both the human protein SelH and the predicted SelH protein conserve the Selenocysteine amino acid.
As we could not correctly predict the protein, the process was repeated using the SelH cow protein. Six hits were obtained when running tblastn, all of them again in the scaffold SJYR01002415.1. The protein was predicted using the first hit (which had an e-value of 5.93e-12) and the number of exons obtained (six exons) is the same as the one obtained when using the human SelH as a query. This protein is located in the positive strand between the positions 99565 and 101372. The tcoffee obtained in the second attempt was better than in the first one (the obtained from the human protein) with an score of 775. We can see that the predicted protein in the second attempt is a little bit larger than the one predicted from the human Selenoprotein, but it still seems that the predicted protein has skipped some exons. It can be observed that both the cow protein SelH and the predicted SelH protein conserve the Selenocyteine amino acid.
Once analyzed all the hits, it seems like there has been several duplications of SelH protein in K. ellipsiprymnus genome. Moreover, it seems SelH protein in K. ellipsiprymnus genome would have lost some exons.
No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein. Taken all together, the protein SelH in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that SelH is a selenoprotein.
Selenoprotein T (with thioredoxin) is a selenoprotein with selenium binding and thioredoxin-disulfide reductase activity because it has a thioredoxin-like fold motif. It is predominantly localized to the ER and Golgi and it is highly expressed during embryonic development [18]. In brain, SelT protects dopaminergic neurons against oxidative stress against cell death and it has been shown that SelT levels are abundantly expressed in neural progenitors essentiality in embryogenesis when neurogenesis is active, but are inappreciable in adult nervous cells. SelT expression is preserved in neuroendocrine cells and in liver cells during the regenerative process. Also, SelT is implicated in ADCYAP1/PACAP-induced calcium mobilization and secretion, modulating the contraction processes and MYLK activation [20]. Taken together, high SelT levels are involved with ontogenesis, tissue maturation and regenerative processes in neural, endocrine and metabolic tissues [21,22].
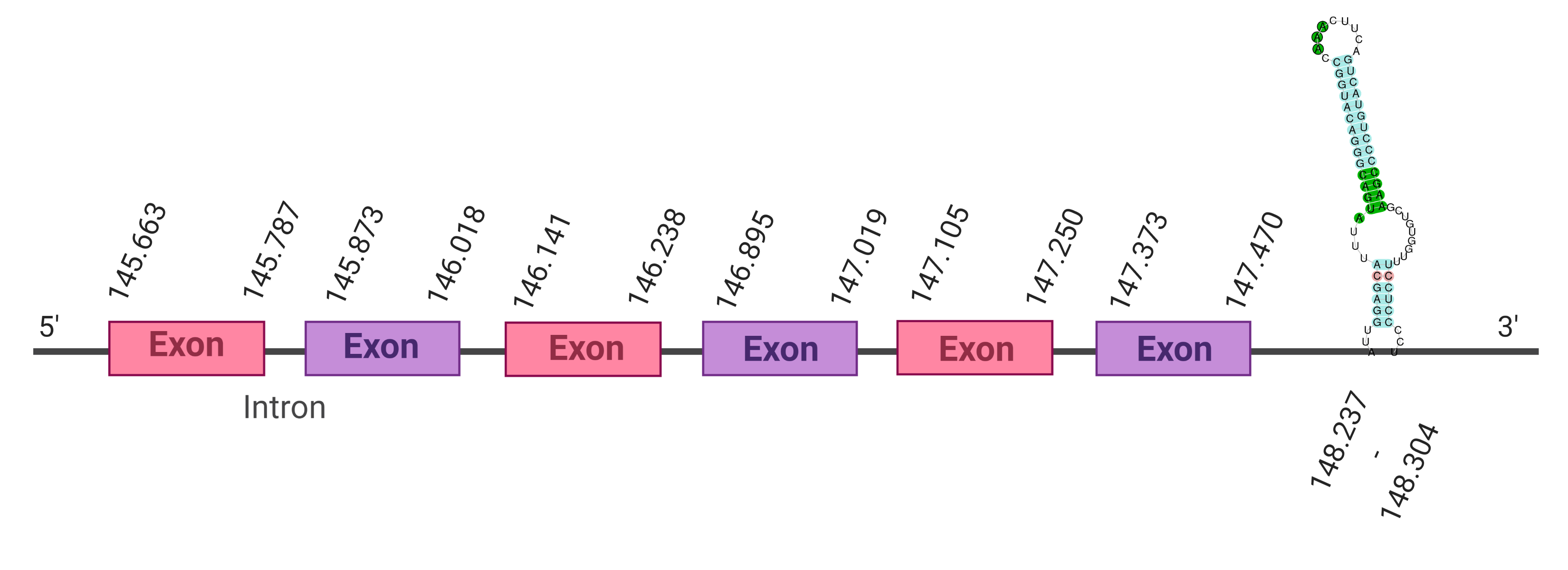
The protein SelT is located in the scaffold SJYR01000122.1 (it was chosen the hit with the lowest e-value: 2.55e-20). Five other hits placed in this same scaffold, in a very similar position, were obtained running tblastn. The protein is located in the negative strand between the nucleotide positions 1600835 and 1626204. In this protein, five exons were predicted. When running tcoffee, it was obtained a score of 997 and visualized a good alignment. It has been predicted correctly all the protein, including the first methionine. It could be observed that both the human protein SelT and the predicted protein in K. ellipsiprymnus conserve the Selenocysteine. No protein was predicted using the Seblastian, but one secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelT in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that SelT is a selenoprotein.
Selenoprotein V (SelV) belong to thioredoxin-like family and was the least characterized mammalian selenoprotein and exclusively found in the placental stem cells. However, it was lost by gene-specific deletion specifically in gorillas. SelV is recently evolved, most likely by a duplication of selenoprotein W (SelW) followed up by the addition of N-terminal sequences - terminal domain, that is not found in SelW [8]. The function of SelV is not known, but may be involved in redox-related processes [22].
Several hits were evaluated for this protein. The first one, the one with the smaller e-value (1e-11), was located in the scaffold SJYR01001274.1. When running tcoffee, it was not possible to align the predicted protein with the genome of interest, obtaining a score of 0.
The second hit, the one with the higher percentage of identity (72%), was located in the scaffold SJYR01000615.1. When running tcoffee,it was obtained the poor score of 770 and visualized a bad alignment with gaps inside the protein sequence.
As the predicted protein obtained from the human protein SelV was not as good as it was expected, SelV protein was predicted again in K. ellipsiprymnus genome using SelV Bos taurus’ protein (Cow). Unfortunately, there’s no such protein for the Bos taurus. It didn’t appear neither in Uniprot nor in Selenodb. Therefore, SelV could not be predicted in K. ellipsiprymnus.
SelW family SelW1
SelW2
SelI
SelI are transmembrane proteins only found in vertebrates. They contain a highly conserved CDP-alcohol phosphatidyltransferase domain, also present in the choline and choline/ethanolamine phosphotransferase. Both enzymes participate in de novo synthesis of some phospholipids. SelI also catalyzes phosphatidylethanolamine biosynthesis, but they can be mainly differentiated from these two mentioned enzymes because SelI contains a Sec residue [2,25]. Nevertheless, apart of the role in the physiological function of the SelI and Sec’s role remains to be elucidated [2].
SelK
SelK proteins are transmembrane proteins located in the ER membrane. These proteins’ expression is upregulated when there is an accumulation of misfolded proteins in the ER. Consequently, SelK may be involved in binding misfolded proteins, targeting them to proteasome-dependent degradation. Moreover, some data suggest that they are also implicated in inflammation and the immune response [2].
SelM
SelM proteins are distant homologs of Sel15 and share 31% sequence identity. As well as Sel15, SelM is a thiol-disulfide oxidoreductase located in the endoplasmic reticulum (ER). However, SelM is mainly expressed in the brain [2]. SelM has been suggested to have a role in leptin signalling and, therefore, it could have an effect on food intake [26].
SelN
Selenoprotein N was one of the first identified selenoproteins through bioinformatic approaches. It has an early role in muscle development and differentiation in zebrafish models. When performing SelN knockout mice, they saw SelN might have an important role in the maintenance of satellite cells, meaning it is required for regeneration of skeletal muscle tissue following injury. In humans, mutations in the SelN gene are associated with a group of disorders named after SEPN1-related myopathies, which are early-onset muscle disorders [2].
SelO
Little is known about SelO. Despite the fact that it has been a decade since it was discovered, no structural nor biochemical characterization of this Selenoprotein has been reported. A wide variety of SelO homologs have been reported among species: bacteria, yeast, animals and plants, but its function is still unknown. However, analysis of vertebrate SelO protein sequences has revealed both a protein kinase domain and the presence of a mitochondrial targeting peptide [2].
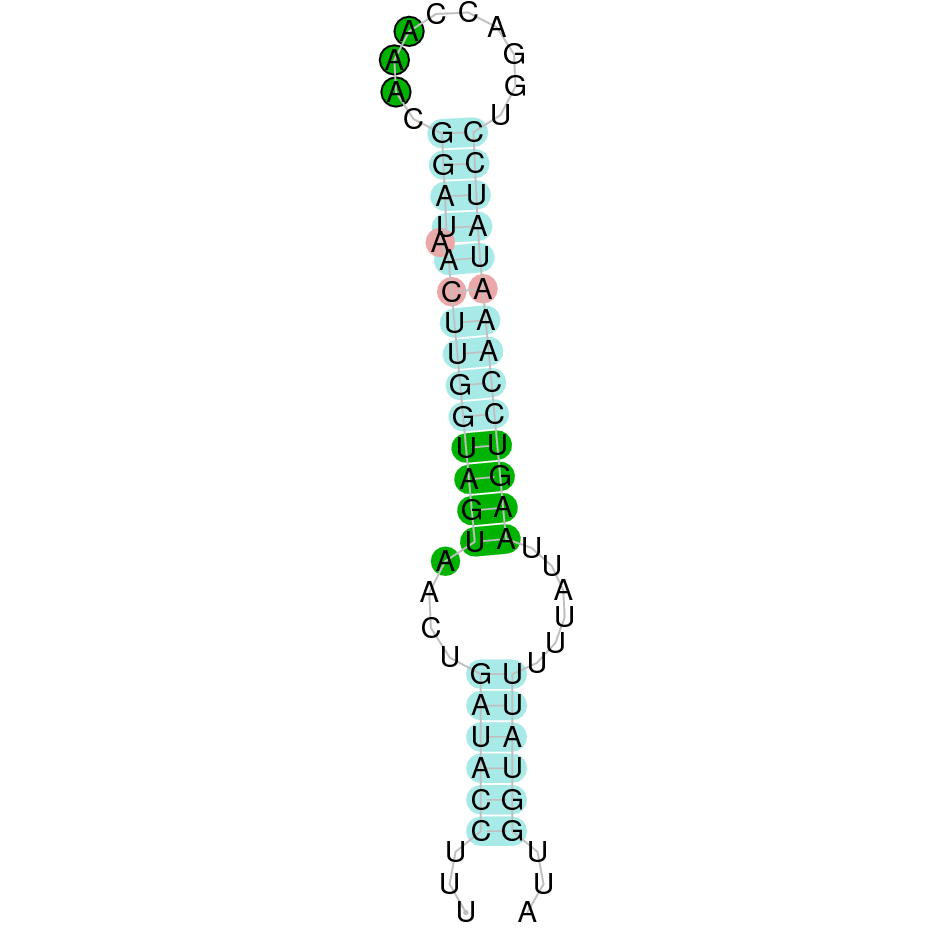
SelP
Selenoprotein P is a highly expressed Seleprotein. It is known that the SelP protein family recently evolved which explains why SelP homologs are found through vertebrates. SelP is involved in transporting Se to peripheral tissue (especially brain and testis). Under extreme conditions of limiting Se, SelP preserves the function of these organs [2].
Selenoprotein W (SelW) is one of the first selenoprotein family characterized and one of the most abundant selenoproteins in mammals. SelW comes from an ancestral selenoproteoma but only SelW1 with the sec-containing form takes part of the selenoproteins found in all the vertebrates. In contrast, SelW2 was lost before the split of amphibians (except in frog) and suffered a Sec-to-Cys conversion generating Rdx12, homolog of SelW2 present in mammals [8]. The function of SelW is not known, but may be involved in a redox-related process, also plays a role in the myopathies of selenium deficiency [23,24].
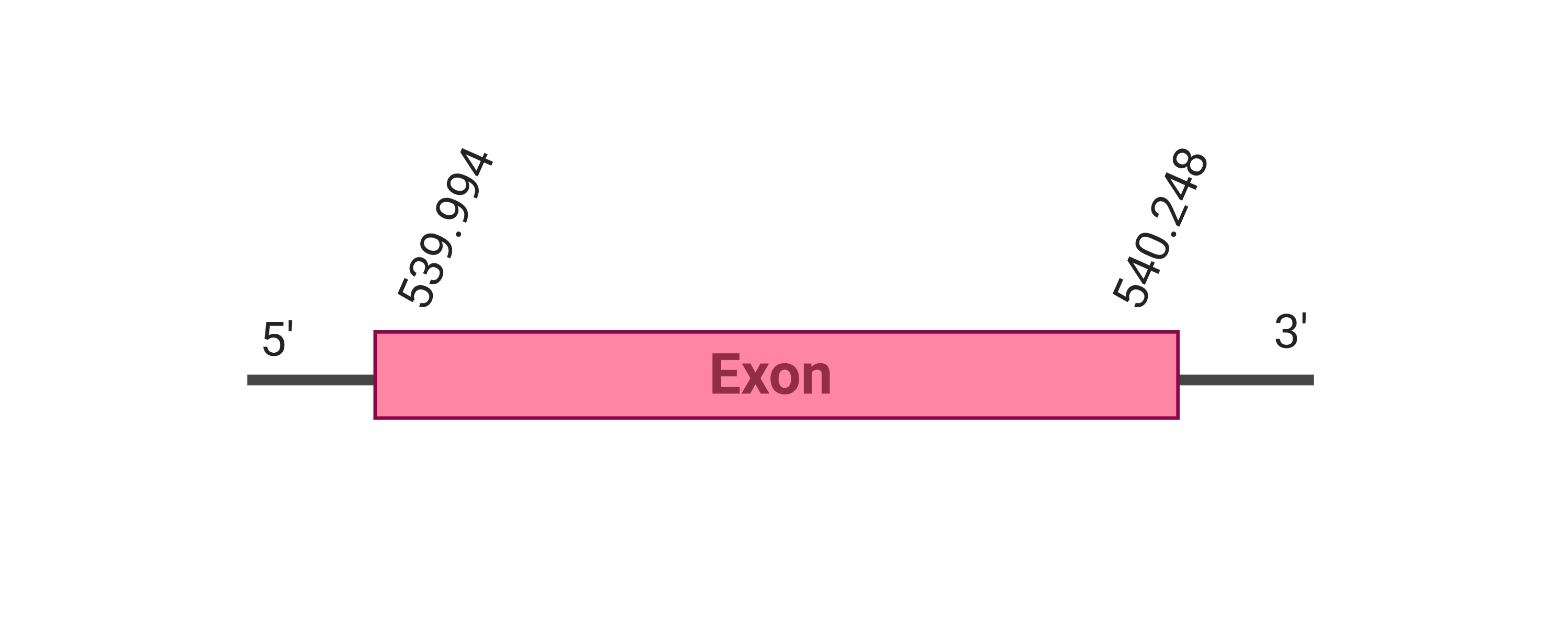
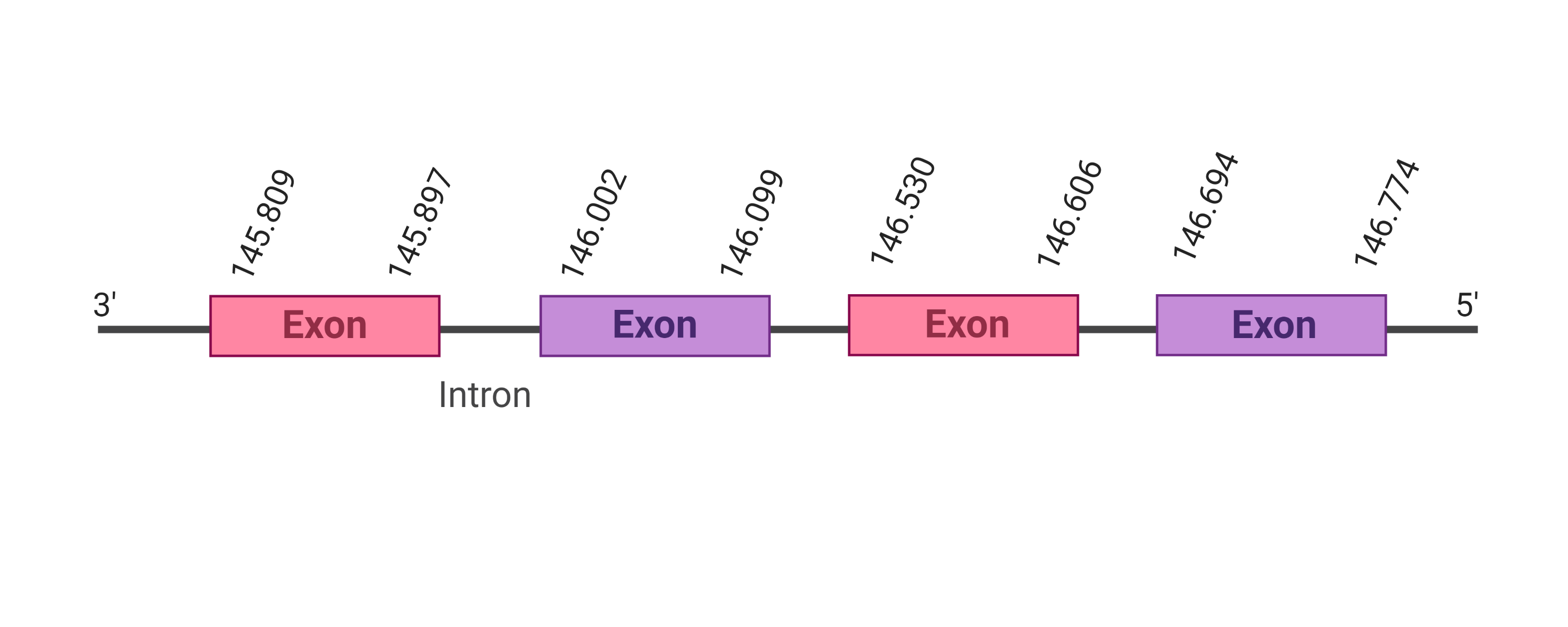
The protein SelW1 is located in the scaffold SJYR01001274.1 (it was chosen the hit with the lowest e-value: 1.00e-21). The percentage of identity between the query and the hit is 82%. The protein is located in the positive strand between the nucleotide positions 539994 and 540248. In this protein, one exon was predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that while the human protein SelW1 conserves the Selenocysteine, the predicted protein in K. ellipsiprymnus has lost it (has an arginine instead).The predicted protein lacks the methionine amino acid at the beginning, meaning that this part of the protein has not been correctly predicted. No protein was predicted using the Seblastian nor any secis element was found in the predicted protein. Additionally, the predicted protein lacks the selenocysteine amino acid. All these signs would suggest that SelW1 is not a selenoprotein.
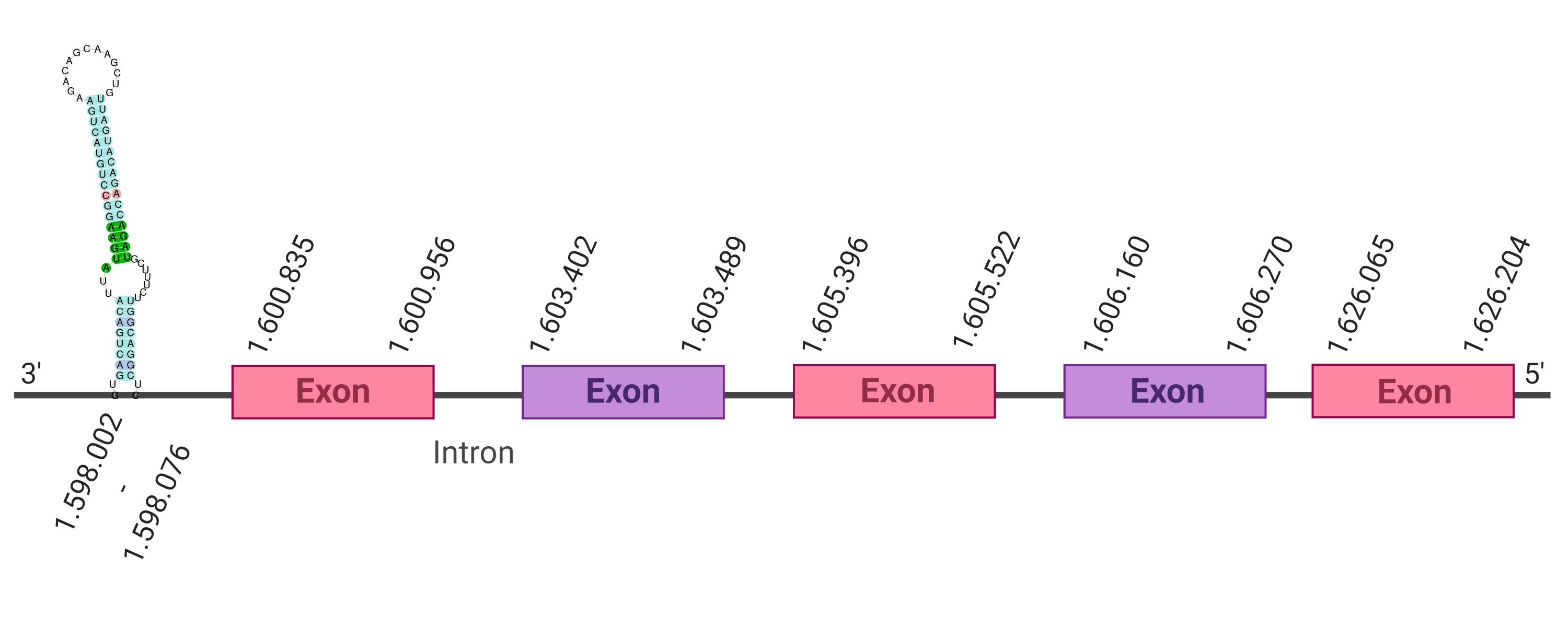
The protein SelW2 is located in the scaffold SJYR01003240.1 (it was chosen the hit with the lowest e-value: 5.51e-21). The percentage of identity between the query and the hit is 63%. The protein is located in the positive strand between the nucleotide positions 145809 and 146774. In this protein, four exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human with the predicted protein and it could be observed that both the human protein SelW3 and the predicted protein in K. ellipsiprymnus do not conserve Selenocysteine. Instead they have a cysteine. The predicted protein starts with a methionine, which means that the beginning of the protein has been correctly predicted. No protein was predicted using the Seblastian nor any secis element was found in the predicted protein. All these signs would suggest that SelW2 is not a selenoprotein.
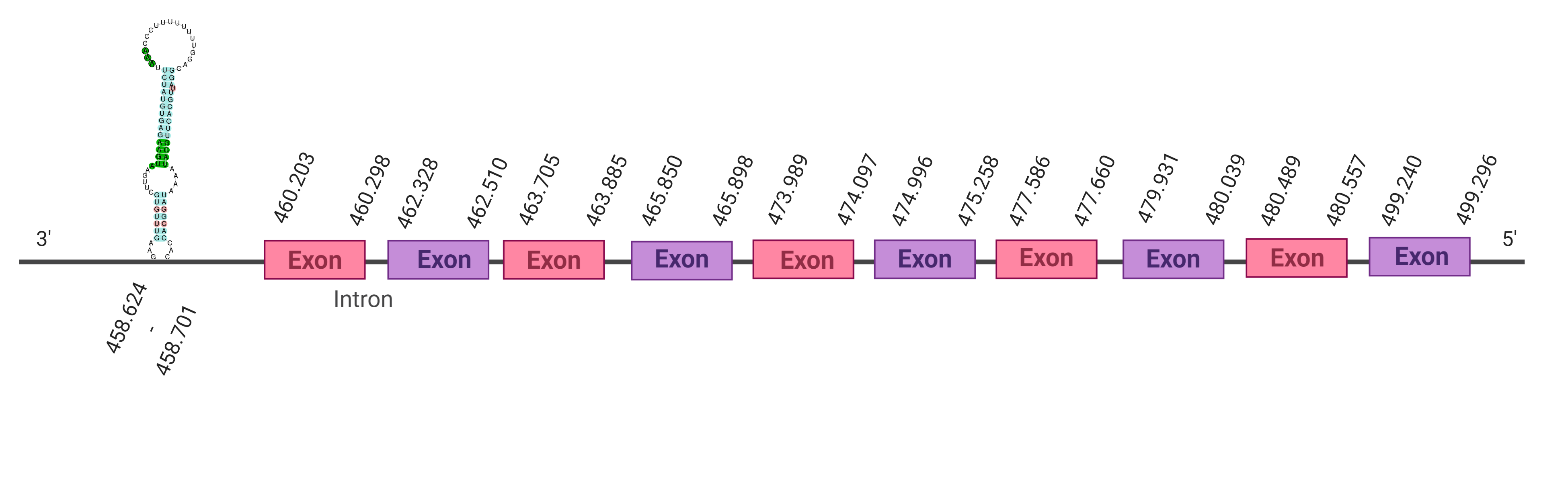
Twelve hits were obtained when running tblastn; three of them were placed in the scaffold SJYR01000772.1 and had the same e-value of 9.90e-153 (despite having different percentages of identity). Taking this under consideration, the hit with the highest percentage of identity has chosen (92%). The predicted protein is located in the positive strand. When running tcoffee, it was obtained a score of 971 and visualized a quite bad alignment with gaps inside the protein sequence. Two exons were predicted in this protein.
The other two hits which had the same e-value were run in the program as well. It was obtained the same tcoffee score, and the same number of exons were predicted. All of this suggests that it turns out to be a triplication. The human SelI conserves the selenocysteine on its sequence.
In order to obtain a better prediction of the protein, we evaluated another hit whose e-value was worse than the first one (6.26e-35). The predicted protein was located in the scaffold SJYR01000634.1. More specifically, it was placed in the reverse strand between the nucleotide positions 460203 and 499296. The percentage of identity between the query and the hit is 98%. When running tcoffee we obtained a score of 1000 and visualized a perfect alignment. In this protein ten exons were predicted. The predicted protein starts with methionine, which means that the beginning of the protein has been predicted correctly. The predicted SelI conserves the Selenocysteine amino acid. Using Seblastian, the selenoprotein ethanolaminephosphotransferase 1 from Bos mutus was predicted in K. ellipsiprymnus. This protein has five exons, and it is located in the negative strand. One Secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelI in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that SelI is a selenoprotein.
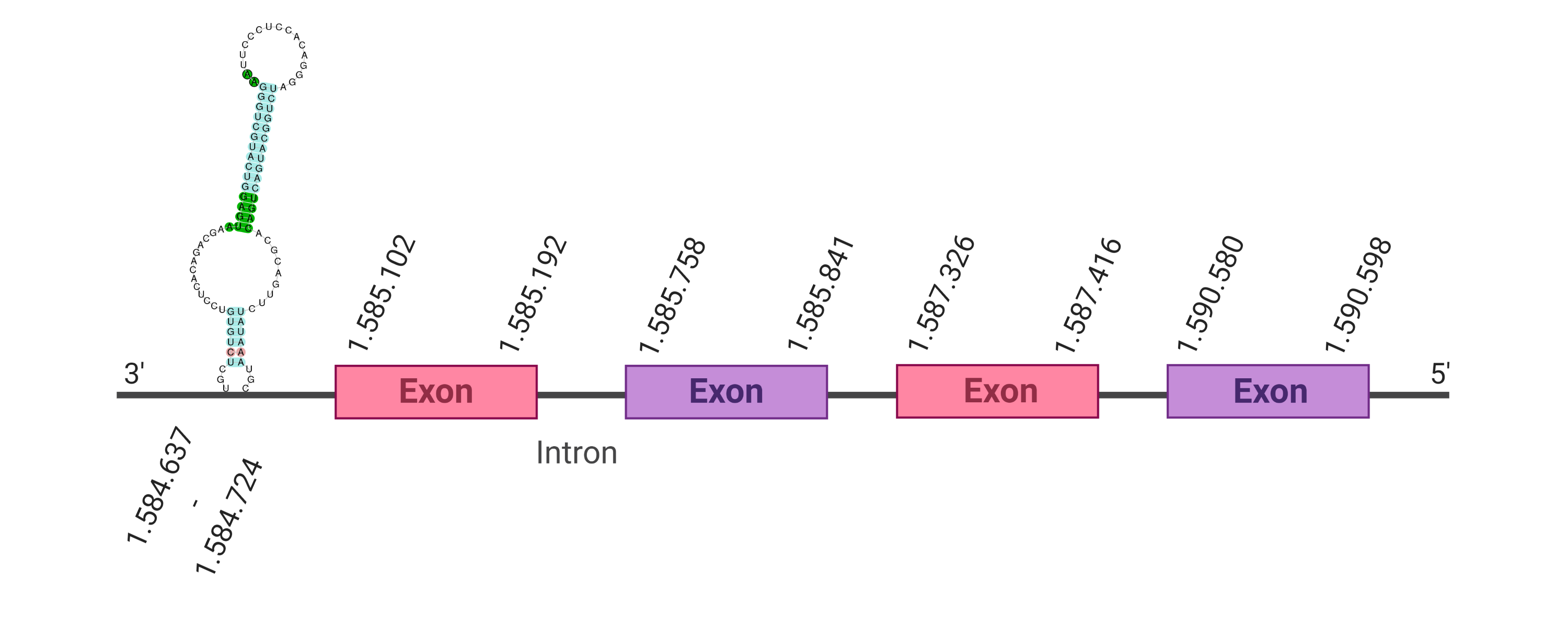
Several hits were evaluated for this protein. The first one, the one with the lowest e-value (1e-20), was located in the scaffold SJYR01000223.1. When running tcoffee, it was obtained a score of 967 and visualized a bad alignment with gaps inside the protein sequence. As the human protein could not be correctly predicted in the genome of our animal, the second hit of the tblastn was evaluated.
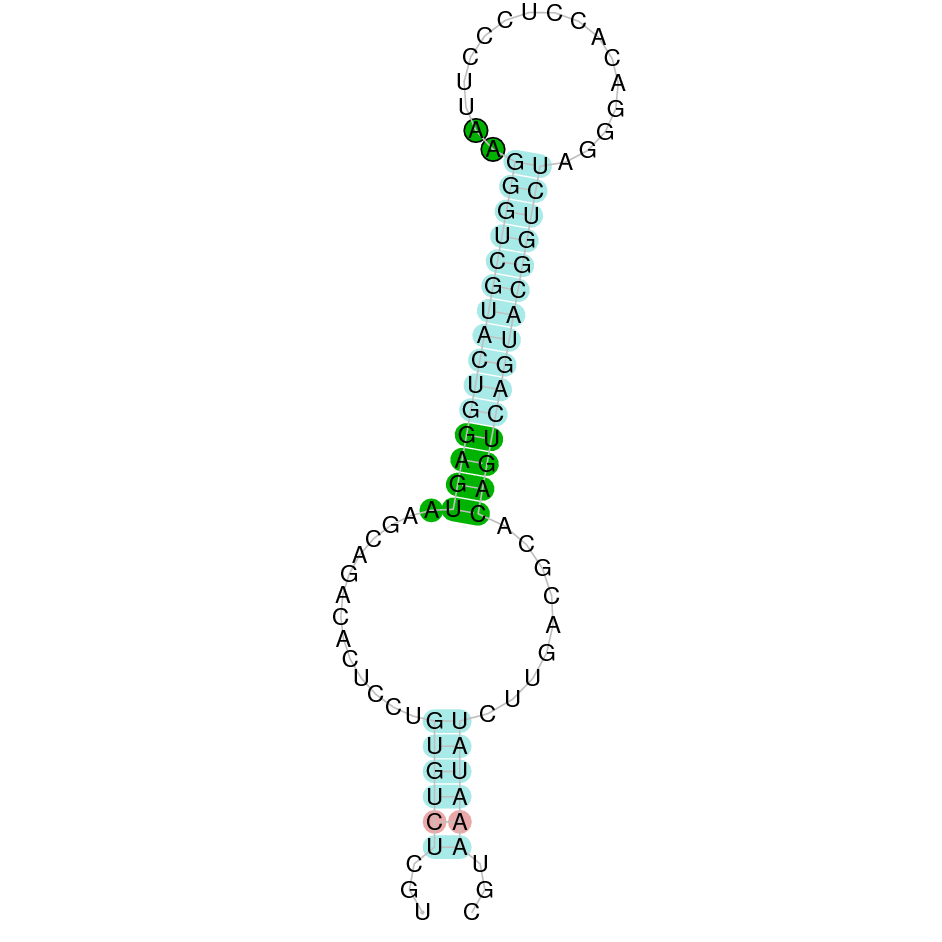
The protein SelK is located in the scaffold SJYR01000121.1 (it was chosen the hit with the lowest e-value: 3.64e-12). The percentage of identity between the query and the hit is 88%. The protein is located in the negative strand between the nucleotide positions 1585102 and 1590598. In this protein, four exons were predicted. When running tcoffee, it was obtained a score of 993 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that both the human SelK protein and the predicted SelK protein in K. ellipsiprymnus conserve the Selenocysteine amino acid. The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. No protein was predicted using the Seblastian, but one secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelK in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that SelK is a selenoprotein.
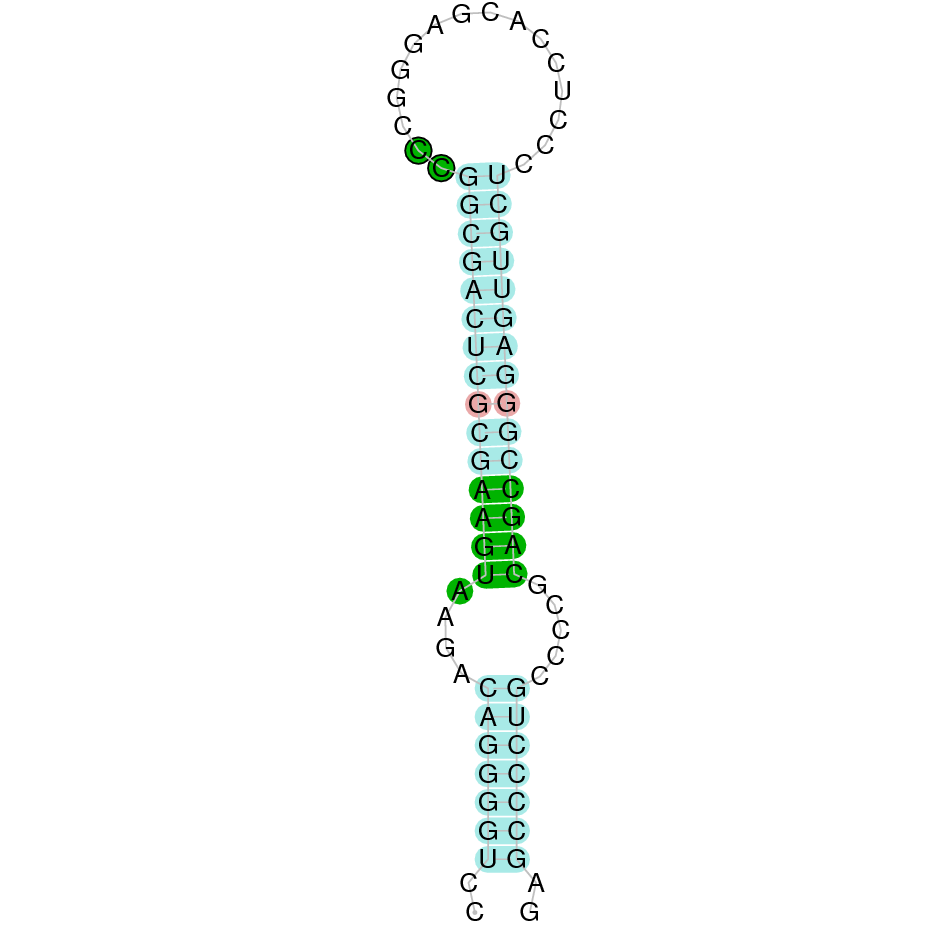
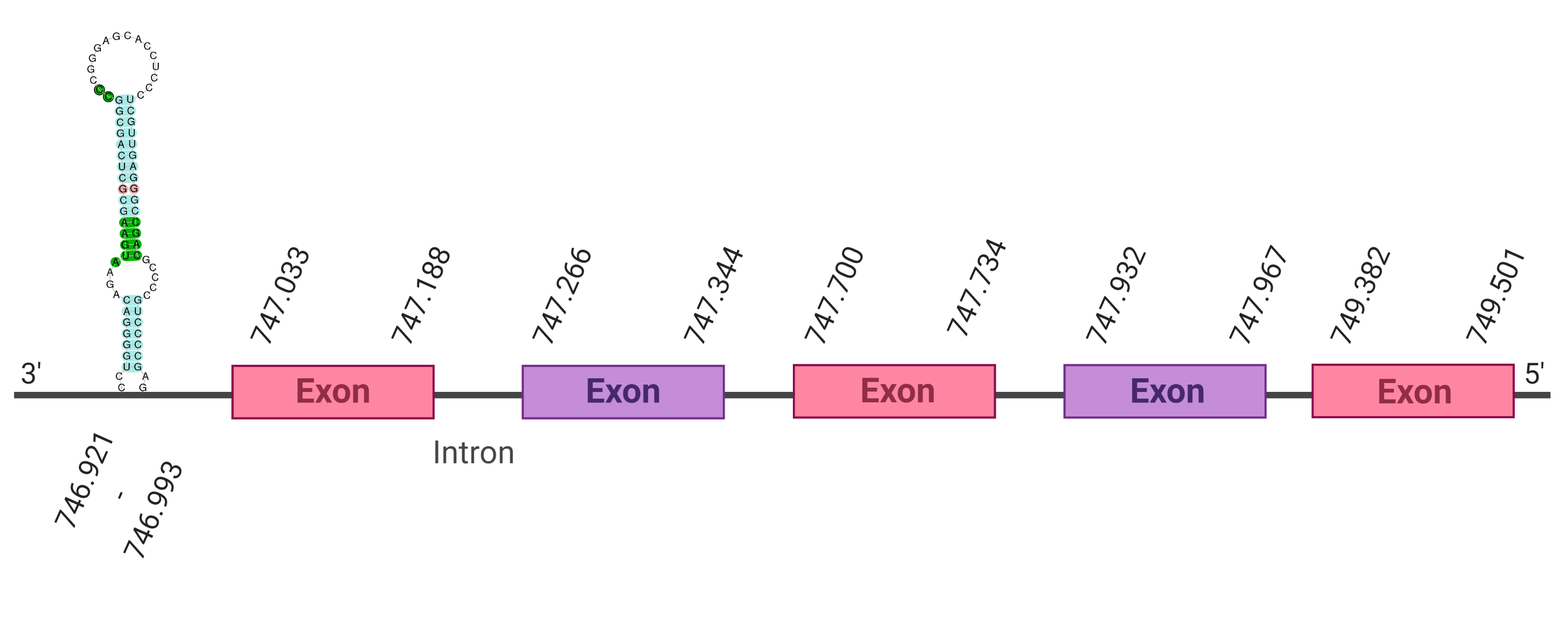
The protein SelM is located in the scaffold SJYR01000283.1 (it was chosen the hit with the lowest e-value: 3.30e-25). When running tcoffee, it was obtained a score of 989, but some gaps inside protein were observed. When looking at the rest of the hits obtained in the tblastn, a quadruplication in the same scaffold was observed. The same results were obtained when the other hits were evaluated. Therefore, SelM protein in K. ellipsiprymnus was predicted again from the SelM Bos taurus’ protein (Cow).
By doing so, the protein SelM is located in the scaffold SJYR01000283.1 (it was chosen the hit with the lowest e-value: 7.59e-25). The percentage of identity between the query and the hit is 97%. The protein is located in the negative strand between the nucleotide positions 747033 and 749501. In this protein, five exons were predicted. The protein, as it had happened before, seems to be quadruplicated in this scaffold. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. Both the cow protein SelM and the predicted SelM protein in K. ellipsiprymnus conserve the Selenocysteine (the human protein SelM conserve as well the Selenocysteine amino acid). The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. No protein was predicted using the Seblastian, but one secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelM in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that SelM is a selenoprotein, that seems to be quatriplicated.
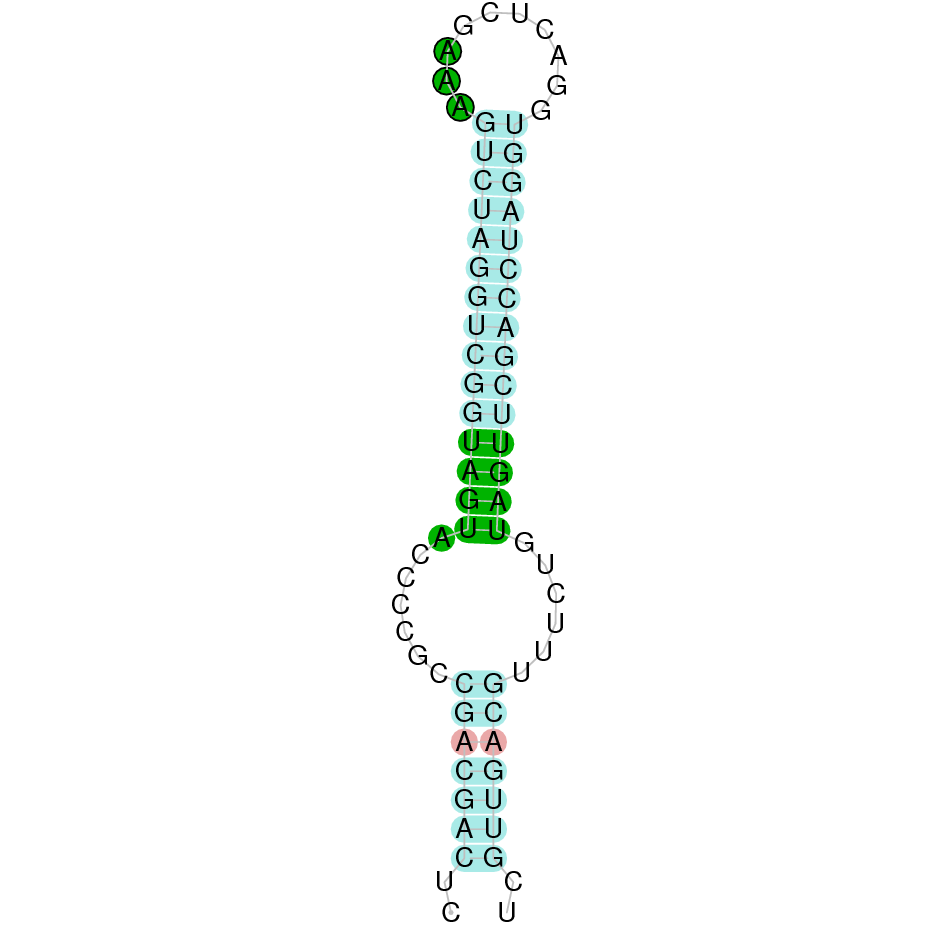
Several hits were evaluated for this protein. The first one was located in the scaffold SJYR01036895.1 (it was chosen the hit with the lowest e-value: 2.05e-31). When running tcoffee, despite having obtained a score of 997, a bad alignment with gaps at the beginning and the end of the protein was given.
As we could not correctly predict the human protein in the genome of our animal, it was evaluated the second hit. The second one was located in the scaffold SJYR01001322.1, had an e-value of 5.87e-30, and its percentage of identity was 90%. The predicted protein is located in the negative strand, between the nucleotide positions 3199 and 20789. In this protein, eleven exons were predicted. When running tcoffee, it was obtained a score of 999. Despite the good score obtained, the first part of the protein could not be predicted (we did not predict the first methionine). The human selenoprotein contained two selenocysteines, while the predicted protein has one selenocysteine (the first one) and one Alanine (the second one, so it is not conserved). No protein was predicted using the Seblastian, but one secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelN in K. ellipsiprymnus contains one selenocysteine and a Secis element. All these signs would suggest that SelN is a selenoprotein.
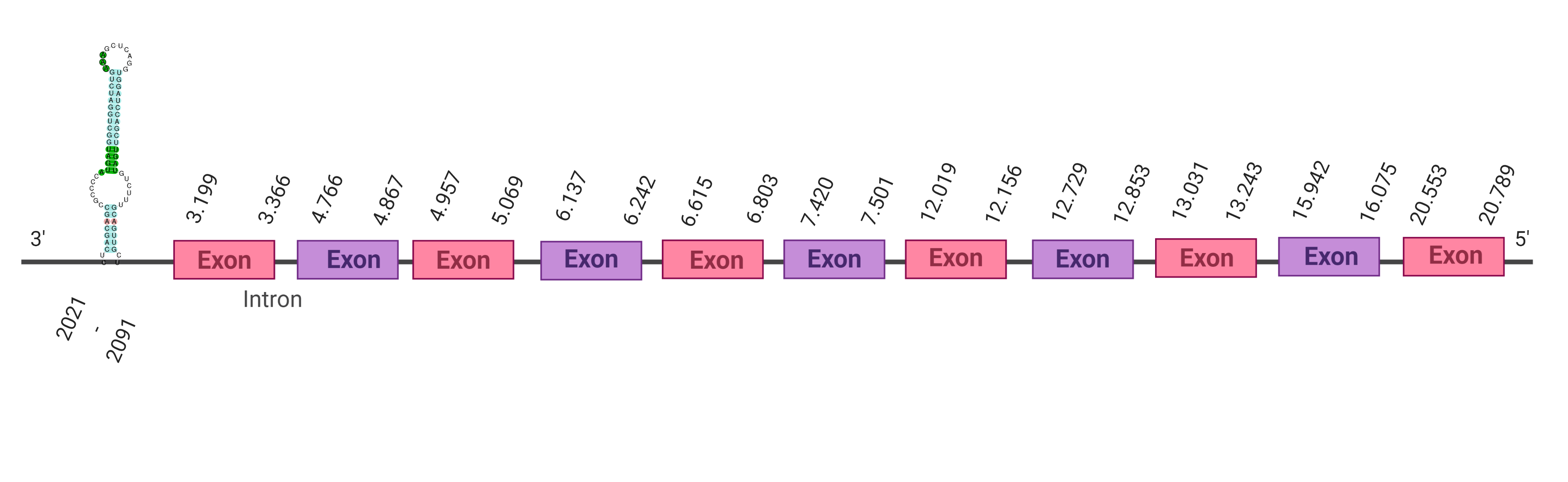
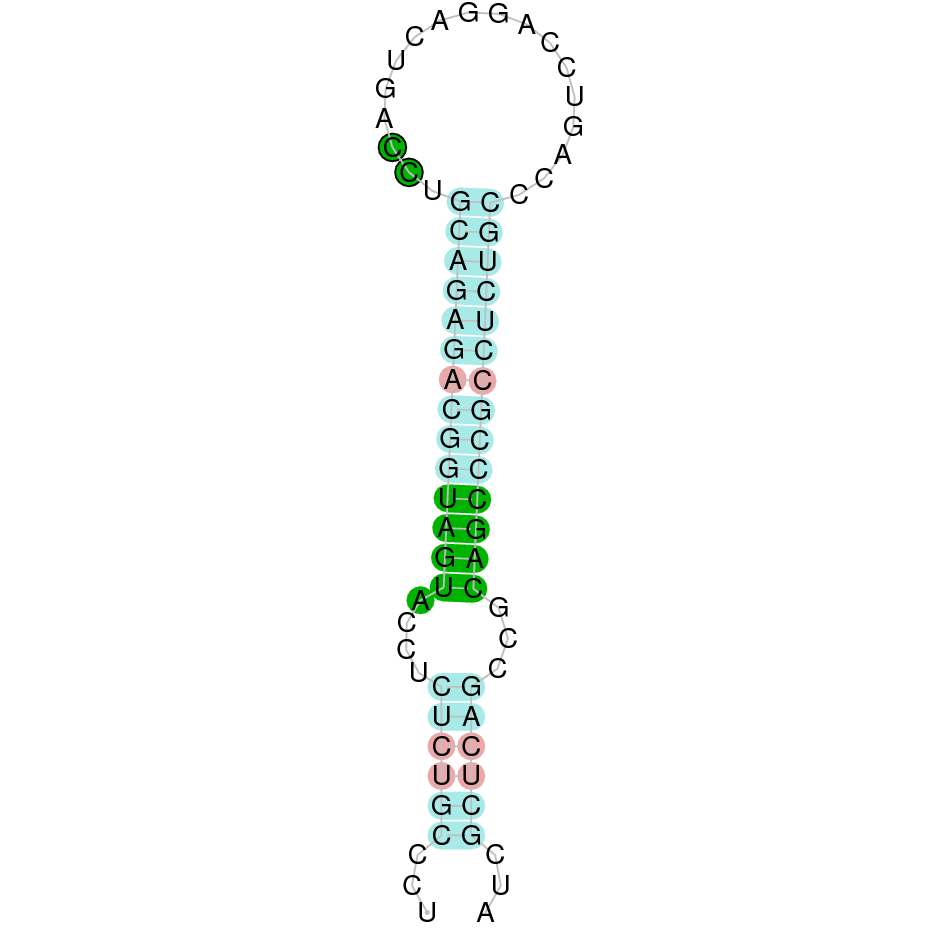
The protein SelO is located in the scaffold SJYR01002254.1 (it was chosen the hit with the lowest e-value: 1.39e-41). The percentage of identity between the query and the hit is 82%. The protein is located in the positive strand between the nucleotide positions 325440 and 344530. In this protein, nine exons were predicted. When running tcoffee, it was obtained a score of 999. Our predicted protein lacks the first methionine amino acid, meaning that the beginning of the protein has not been correctly predicted. Additionally, the final part of the protein could not also be predicted and, it is just in this part where the selenocysteine amino acid is present in the human SelO protein. Therefore, it cannot be concluded if the selenocysteine amino acid is conserved.
Using Seblastian, the selenoprotein O (isoform X2) from Ovis aries musimon was predicted in K. ellipsiprymnus. This protein has three exons, and it is located in the positive strand. Also, one Secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the predicted protein SelO in K. ellipsiprymnus cannot be evaluated whether it contains or not selenocysteine (this region has not been correctly predicted), but it contains a Secis element. As the selenocysteine amino acid was not present in the predicted protein, the results suggest that SelO is not a selenoprotein.
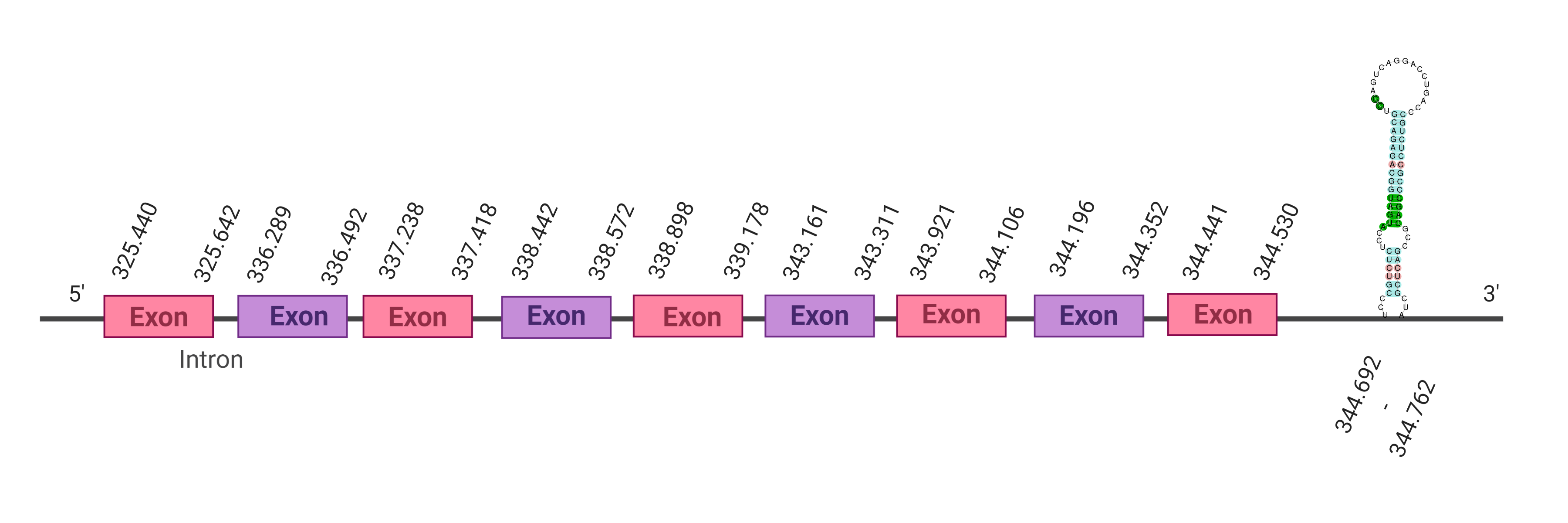
Several hits were evaluated for this protein. The first one, the one with the lowest e-value (1e-32), was located in the scaffold SJYR01002566.1. When running tcoffee, it was obtained the score of 980 and visualized a bad alignment with gaps inside the protein sequence. It seemed that the predicted protein from human lacks one or more exons. When the alignment with the second hit of the tblastn was evaluated, the same tcoffee score was obtained.
As the complete protein sequence of SelP could not be predicted, the SelP from Bos taurus was used as a query. The first hit was located in the scaffold SJYR01002566.1 (which had an e-value of 4.72e-48). The predicted protein is placed in the negative strand between the nucleotide positions 365231 and 359708. The predicted protein has four exons. When running tcoffee, it was obtained a score of 985, and it could be observed that, as it had happened with the human protein, it was predicted the first (including the methionine) and the last part of the protein. It also seems that the predicted Kobus ellipsiprymnus protein had lost some exons when predicted both from the cow and the human Selenoprotein P. The predicted protein in our animal conserves the selenocysteine present in both the human and cow SelP.
Using Seblastian, the selenoprotein P from Capra hircus was predicted in K. ellipsiprymnus. This protein has one exon, and it is located in the negative strand. Also, one Secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelP in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that SelP is a selenoprotein.
Methionine-R-sulfoxide reductase family
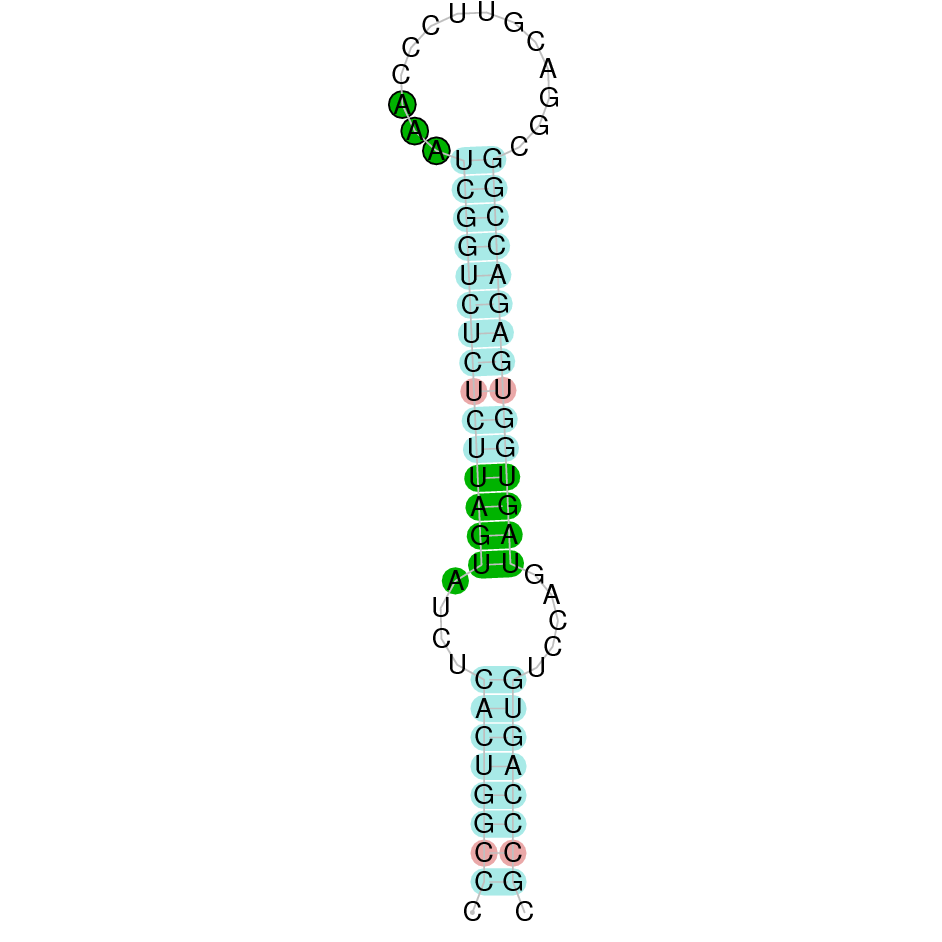
SelR1 (also known as Methionine-R-sulfoxide reductase 1 or MsrB1) is involved in reducing methionine (R)-sulfoxide back to methionine. Methionine oxidation is a type of post-translational modification. SelR1 has a role in regulating actin assembly and promoting filament repolymerization [27].
SelR2 (Methionine-R-sulfoxide reductase 2 or MsrB2) also reduces methionine (R)-sulfoxide back to methionine. On the other hand, this subtype of Selenoprotein is involved in preserving mitochondrial integrity by decreasing the intracellular reactive oxygen species, watching after cell survival [28].
Finally, SelR3 (Methionine-R-sulfoxide reductase 3 or MsrB3) also reduces methionine (R)-sulfoxide back to methionine. Concerning its sequence, it contains both a signal peptide and a mitochondrial transit peptide [29].
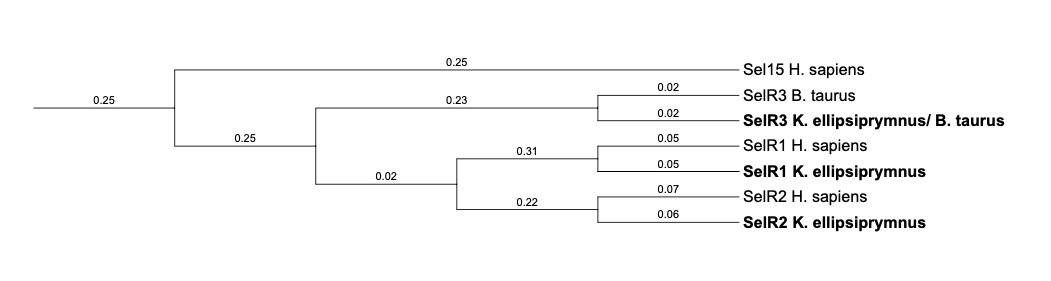
In the phylogenetic tree of Methionine-R-sulfoxide reductase family, it can be seen that the selenoproteins SelR1, SelR2 and SelR3 predicted in K. ellipsiprymnus are classified together with their own queries. It suggests that the proteins have been predicted correctly.
SelR1
SelR2
SelR3
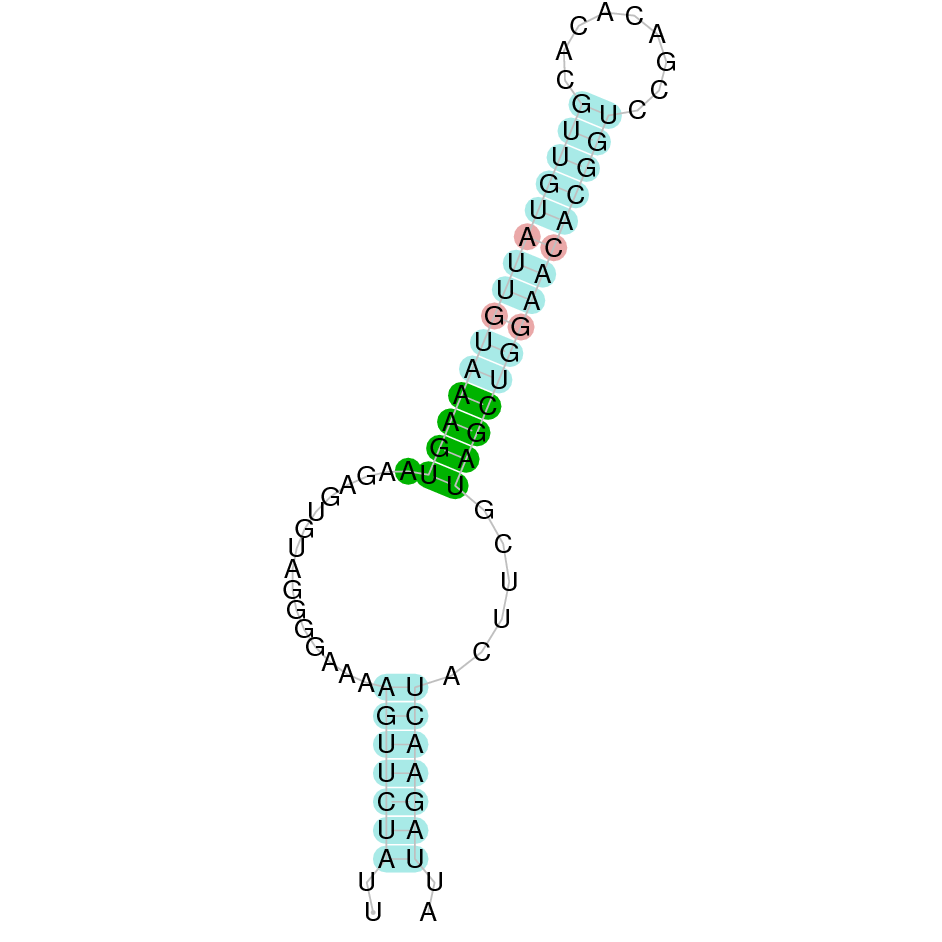
SelS
Selenoprotein S has a role in the degradation of misfolded endoplasmic reticulum (ER) luminal proteins. In particular, it guides misfolded protein from the ER to the cytosol. Once there, proteins are destroyed by the proteasome. The hypothesis says that it might act as a linker between DERL1 (which is in charge of the retrotranslocation of the misfolded proteins into the cytosol) and the ATPase complex VCP (in charge of mediating both the translocation and ubiquitination) [30].
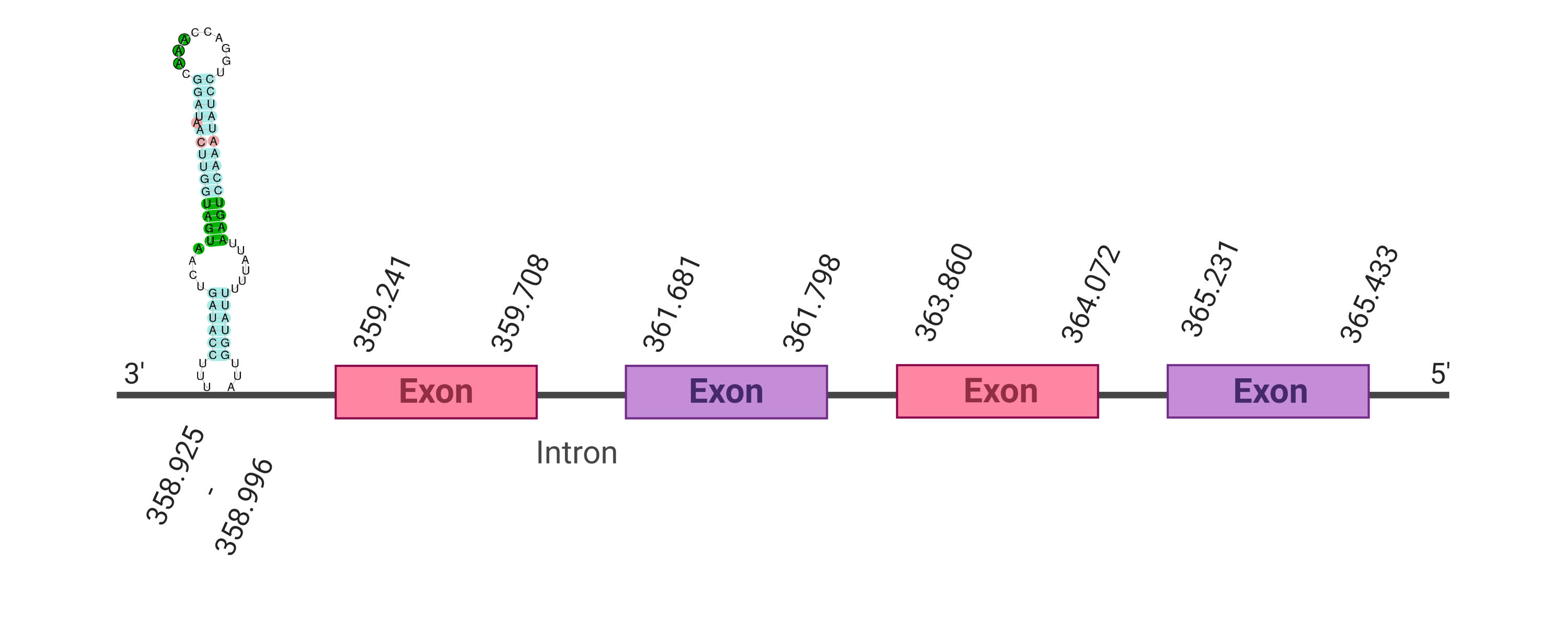
Four hits were obtained when running tblastn, all of them placed in the scaffold SJYR01001528.1. The protein was predicted using the first hit that had the lowest e-value (5.48e-24) and a percentage of identity of 68%. The predicted protein is located in the reverse strand between the nucleotide positions 189017 and 193966 and contains four exons. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. The predicted protein starts with methionine, which means that the beginning of the protein has been correctly predicted. Tcoffee aligns the human protein with the predicted protein and, it can be observed that both of them conserve the selenocysteine amino acid. Using Seblastian, the selenoprotein methionine-R-sulfoxide reductase B1 from Ovis aries was predicted in K. ellipsiprymnus. This protein has three exons, and it is located in the negative strand. Also, one Secis element (grade A) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelR1 in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that SelR1 is a selenoprotein.
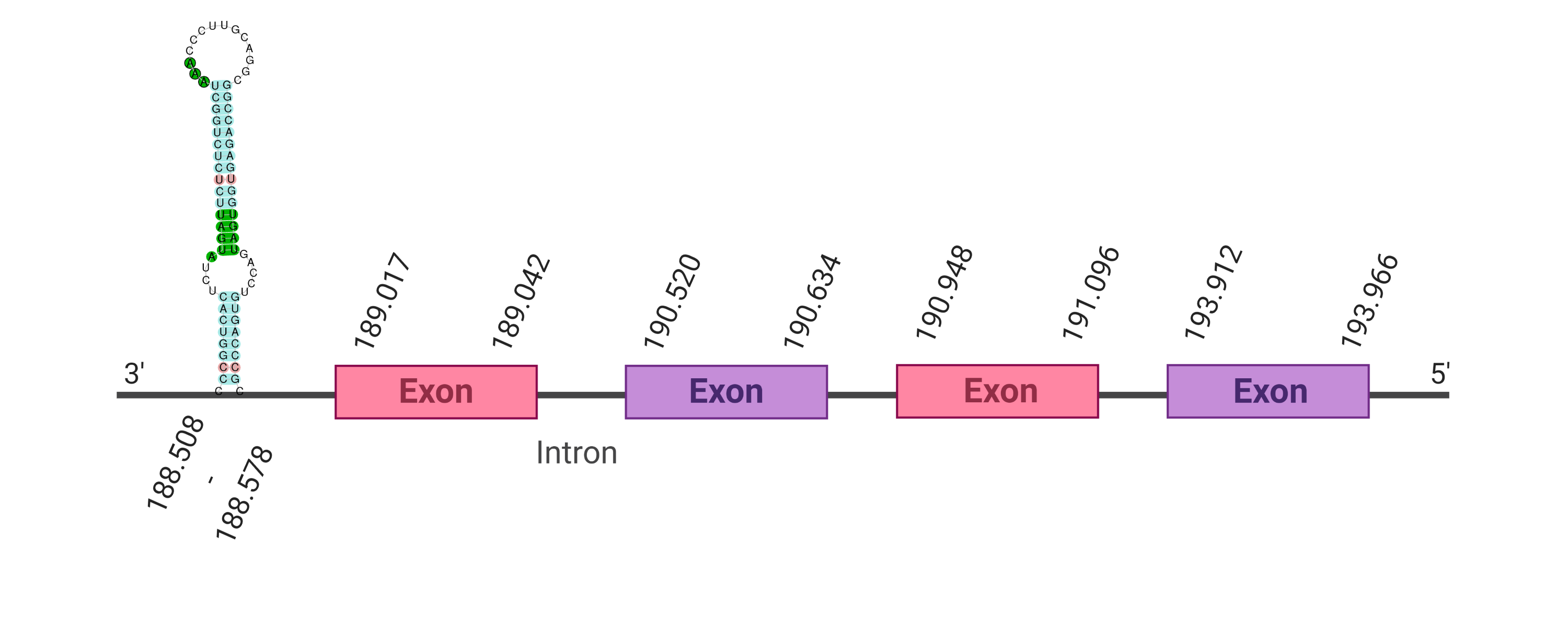
The protein SelR2 is located in the scaffold SJYR01000378.1 (it was chosen the hit with the lowest e-value: 3.36e-21). When running tcoffee, it was obtained a score of 999. Despite having obtained such a good score, the first part of the protein was not correctly predicted.
In order to obtain a better prediction of the protein, it was evaluated the second hit whose e-value is 4.69e-10. The protein SelR2 is located in the scaffold SJYR01000378.1. The percentage of identity between the query and the hit is 93%. The protein is located in the negative strand between the nucleotide positions 103402 and 117923. In this protein, four exons were predicted. When running tcoffee, it was obtained a score of 999. Although most of the region is correctly aligned, the first part of the protein was not reached to be aligned. It can be observed that both the human protein SelR2 and the predicted SBP2 protein in K. ellipsiprymnus did not conserve selenocysteine (instead they have a cysteine). The predicted protein lacks the first methionine amino acid, meaning that the beginning of the protein sequence has not been predicted. Any selenoprotein was predicted using the Seblastian, nor any Secis element was found in the predicted protein. This suggests that nowadays, K. ellipsiprymnus’ SelR2 is a cysteine-homologous protein.
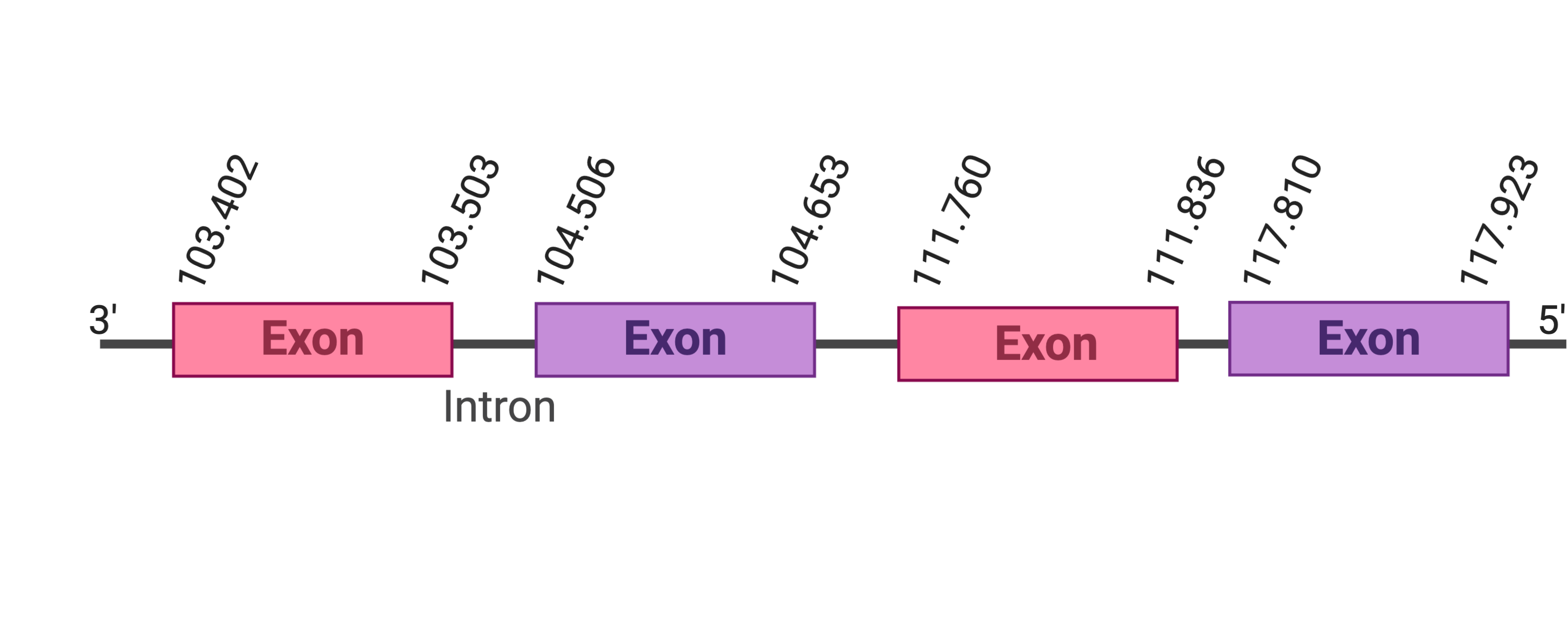
Several hits were evaluated for this protein. The first hit was located in the scaffold SJYR01001010.1 (it was chosen the hit with the lowest e-value: 2.23e-18). When running tcoffee, it was obtained a score of 972 and it could be observed that it had only been predicted the last part of the protein.
In order to obtain a better predicted protein, the SelR3 cow selenoprotein was used as a query. Firstly, it was evaluated the first hit (the one with the lowest e-value 1.42e-14), which was placed in the scaffold SJYR01000020.1. The protein is located in the positive strand between the nucleotide positions 699131 and 765343. Three exons were predicted in this protein. When running tcoffee, it was obtained a score of 1000. Our predicted protein, despite having obtained such a good score, only contains the first part of the protein (including the methionine). Both the cow protein and the predicted protein do not contained selenocysteine nor cysteine.
As there was a second hit in another scaffold with a similar e-value (3.39e-13), it was evaluated. The predicted protein was placed in the scaffold SJYR01001010.1, in the positive strand between the nucleotide positions 24614 and 106809. The predicted protein has three exons. When running tcoffee, it was obtained a score of 987. Our predicted protein only contains the second part of the protein and, once again, it does not contain any selenocysteine nor cysteine.
If taken into account the two predicted proteins from cow SelR3 protein and the analysis of the two tcoffees obtained, it can be concluded that the first part of the protein is located in the scaffold SJYR01000020.1 and the final part of the protein is located in the scaffold SJYR01001010.1 (therefore, all the protein has been correctly predicted). It can be concluded that the SelR3 protein in Kobus ellipsiprymnus has in total six exons: three in one scaffold and the left three in the other scaffold.
Seblastian predicted the selenoprotein P1 from Capra hircus in the negative strand. The predicted protein only has one exon and, it was discarded as, according to our results, SelR3 is placed in the positive strand. No secis element was found in the predicted protein. This suggests that K. ellipsiprymnus’ SelR3 is not a selenoprotein.
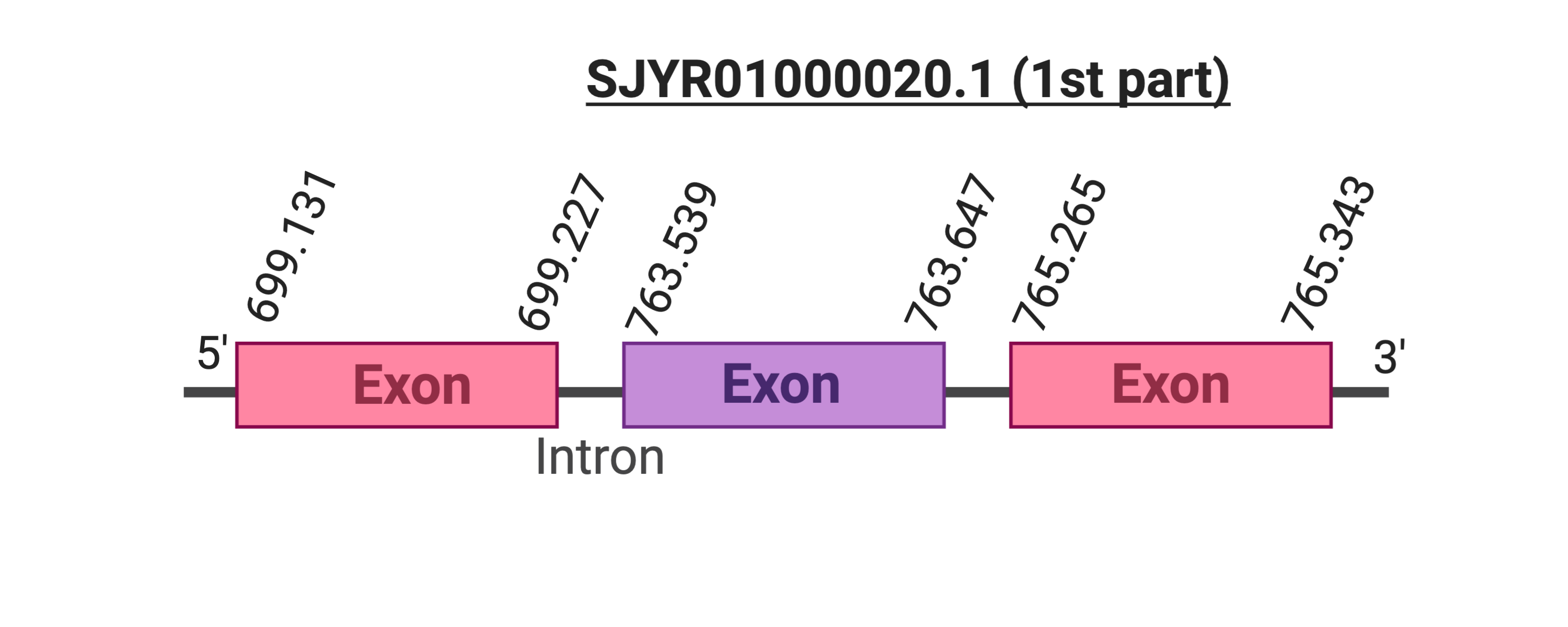
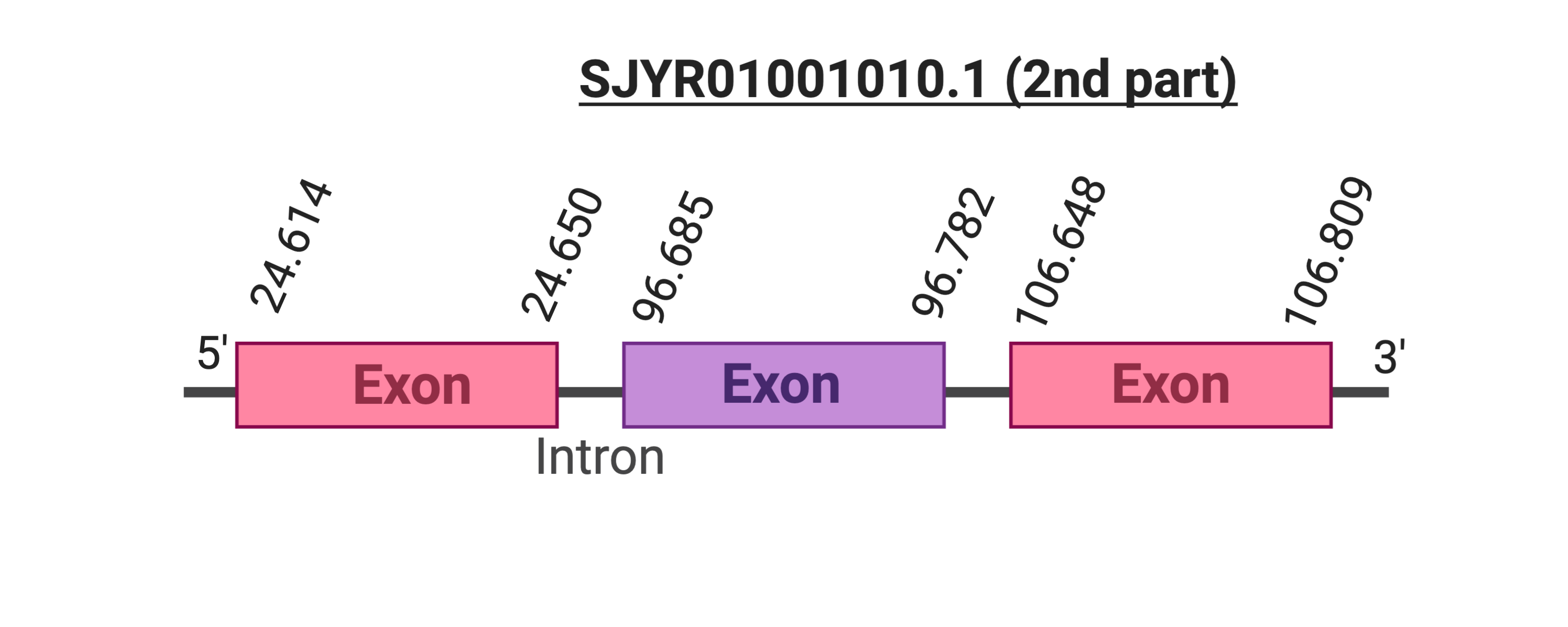
The protein SelS is located in the scaffold SJYR01003258.1 (it was chosen the hit with the lowest e-value: 1.20e-10). Three other hits placed in the same scaffold, in a very similar position, were obtained running tblastn. The protein is located in the negative strand between the nucleotide positions 291560 and 299601. In this protein, six exons were predicted. When running tcoffee, it was obtained a score of 996 and visualized quite a good alignment. It has been predicted correctly all the protein, including the methionine. It could be observed that both the human protein SelS and the predicted protein in K. ellipsiprymnus conserve the Selenocysteine. Any selenoprotein was predicted using the Seblastian, and only one element Secis was found in the positive strand.
The lack of a Secis element in the negative strand suggests that K. ellipsiprymnus’ SelS is not a selenoprotein.
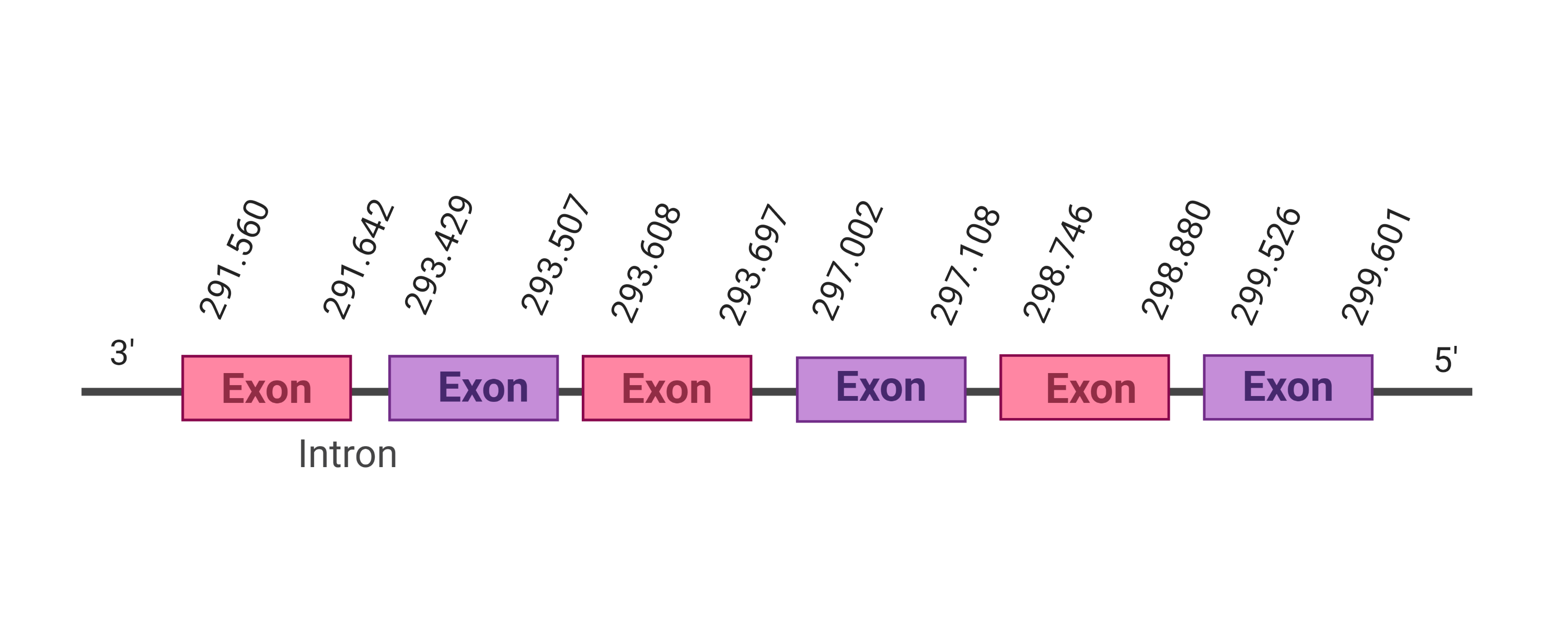
SelU family
SelU family (also known as tRNA 2-selenouridine synthase family) is a newly identified selenoprotein family, that includes SelU1, SelU2 and SelU3 homolog proteins. This SelU family were firstly reported in the puffer fish, although the Sec-containing form also appeared in other fish, sea urchins, diatoms, green algae and chickens. However, mammals, worms and arthropods contained cysteine homologues, the active-site sec was replaced by cysteine. Recent studies shown evidence that supports a Sec-to-Cys conversion from Sec-containing SelU1 sequences in fish to Cys-containing SelU1 proteins in mammals (it could not been proved in homolog proteins SelU2 and SelU3) [8,22].
The function of this family remains unclear, although it is proved that SelU plays a key role for the survival and normal functioning of SCs in chickens due to deprivation of SelU induced autophagy, altered the expression of growth factor secreted by Sertoli cells (SCs), such as IGF1, and inhibited apoptosis via PI3K–Akt pathway [31,32].
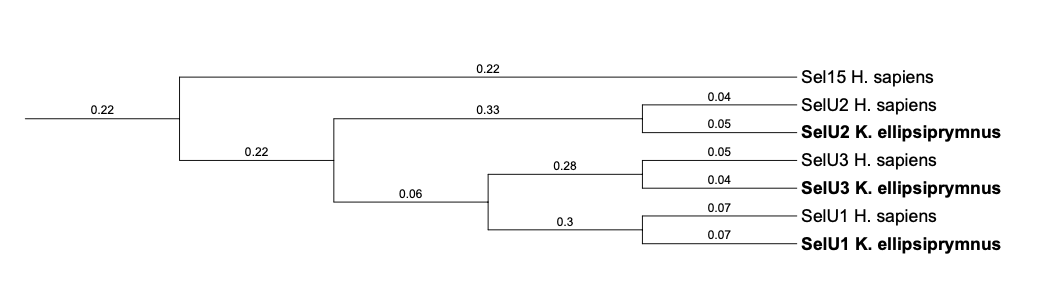
In the phylogenetic tree of SelU family, it can be seen that the selenoproteins SelU1, SelU2 and SelU3 predicted in K. ellipsiprymnus are classified together with their own queries. It suggests that the proteins have been predicted correctly.
SelU1
SelU2
SelU3
The protein SelU1 is located in the scaffold SJYR01001912.1 (it was chosen the hit with the lowest e-value: 8.27e-24). The protein is located in the negative strand between the nucleotide positions 362613 and 366415. In this protein, five exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. It has been predicted correctly all the protein, including the first methionine. It could be observed that both the human protein SelU1 and the predicted protein in K. ellipsiprymnus do not conserve the Selenocysteine, they have a cysteine. No protein was predicted using the Seblastian, but one secis element (grade B) in the negative strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein SelU1 in K. ellipsiprymnus contains cysteine and a Secis element. All these signs would suggest that SelU1 is a cysteine homologous protein.
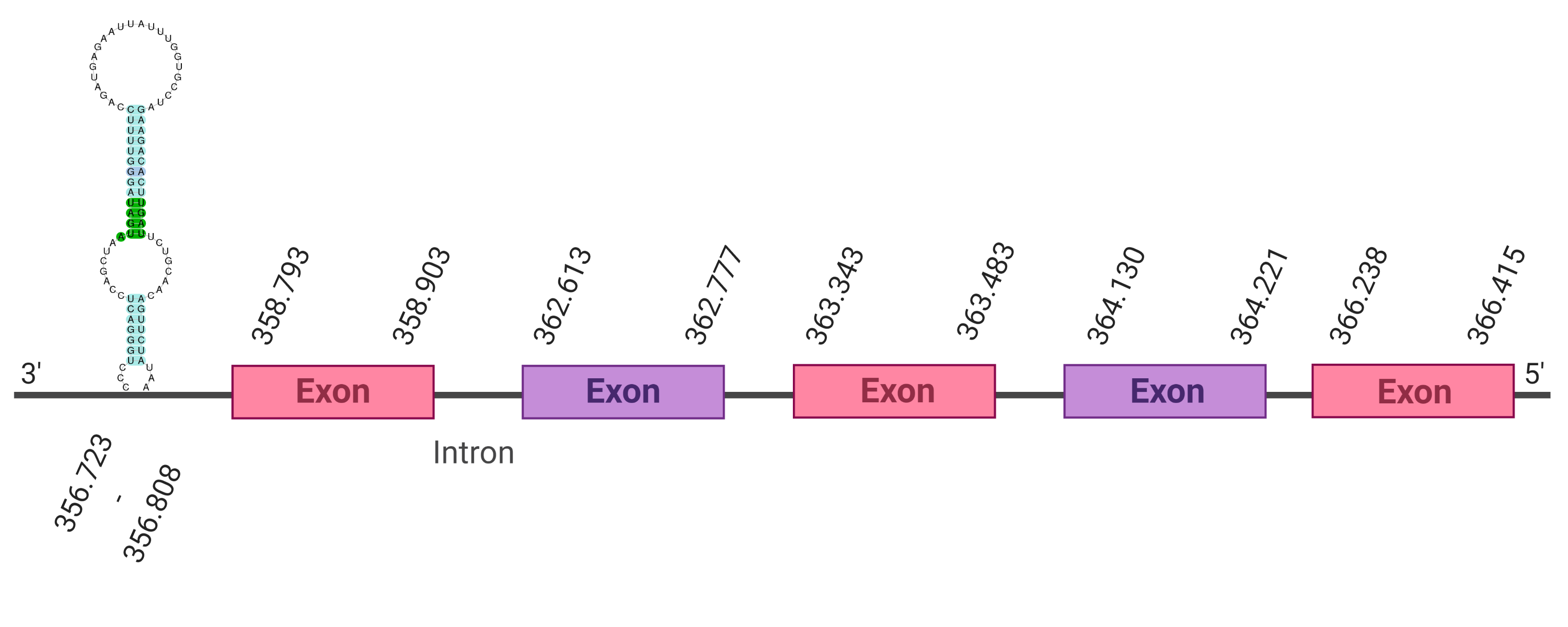
The protein SelU2 is located in the scaffold SJYR01001714.1 (it was chosen the hit with the lowest e-value: 2.18e-19). The protein is located in the positive strand between the nucleotide positions 148779 and 167520. In this protein, six exons were predicted. When running tcoffee, it was obtained a score of 997 and visualized quite a perfect alignment. It has been predicted correctly all the protein but there are some mismatches and it could not be correctly predicted the first methionine (instead, it has been predicted a Serine). It could be observed that both the human protein SelU2 and the predicted protein in K. ellipsiprymnus do not conserve the Selenocysteine, they have a cysteine. No protein was predicted using the Seblastian nor any secis element was found in the predicted protein. All these signs would suggest that SelU2 is a cysteine homologous protein.
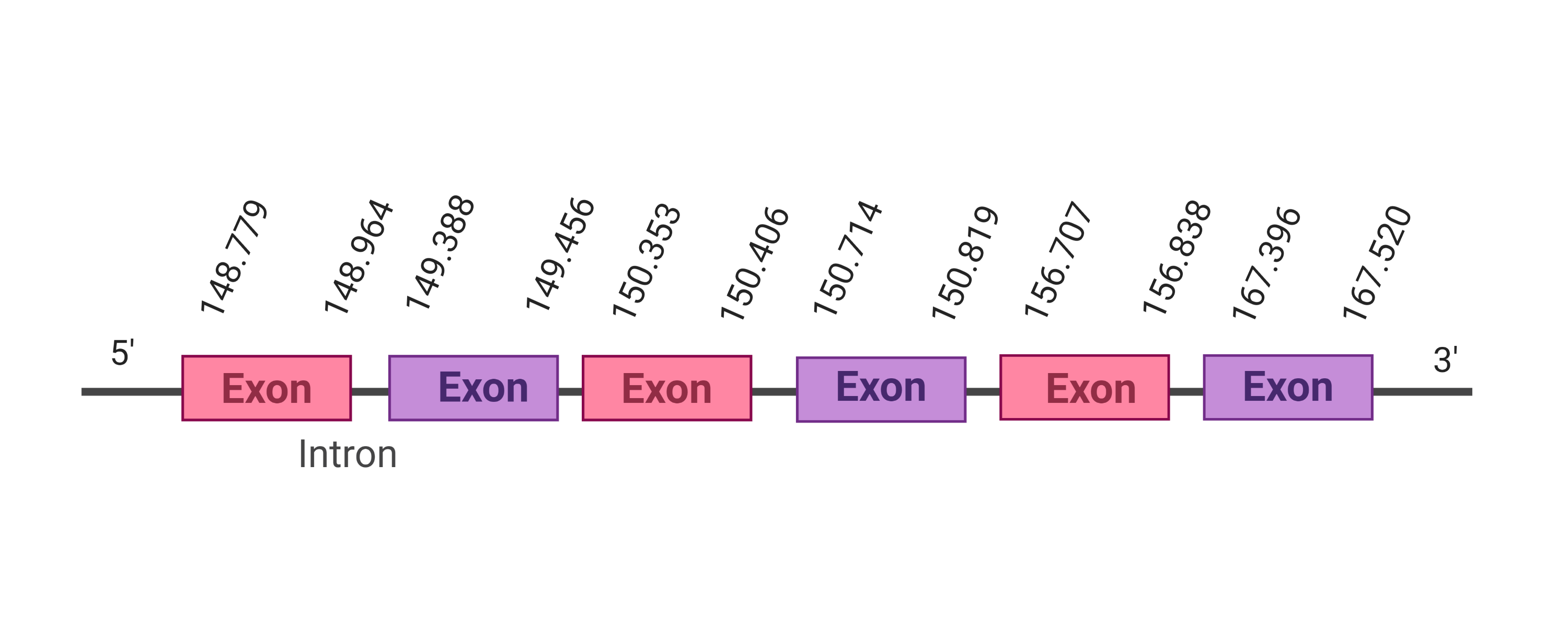
The protein SelU3 is located in the scaffold SJYR01001383.1 (it was chosen the hit with the lowest e-value: 4.76e-31). The protein is located in the negative strand between the nucleotide positions 271344 and 273745. In this protein, six exons were predicted. When running tcoffee, it was obtained a score of 1000 and visualized a perfect alignment. It has been predicted correctly all the protein except for the last four amino acids (it was predicted the first methionine). It can be observed that both the human protein SelU3 and the predicted protein in K. ellipsiprymnus do not conserve the Selenocysteine, and instead they have a cysteine. No protein was predicted using the Seblastian nor any secis element was found in the predicted protein. All these signs would suggest that SelU3 is a cysteine homologous protein.
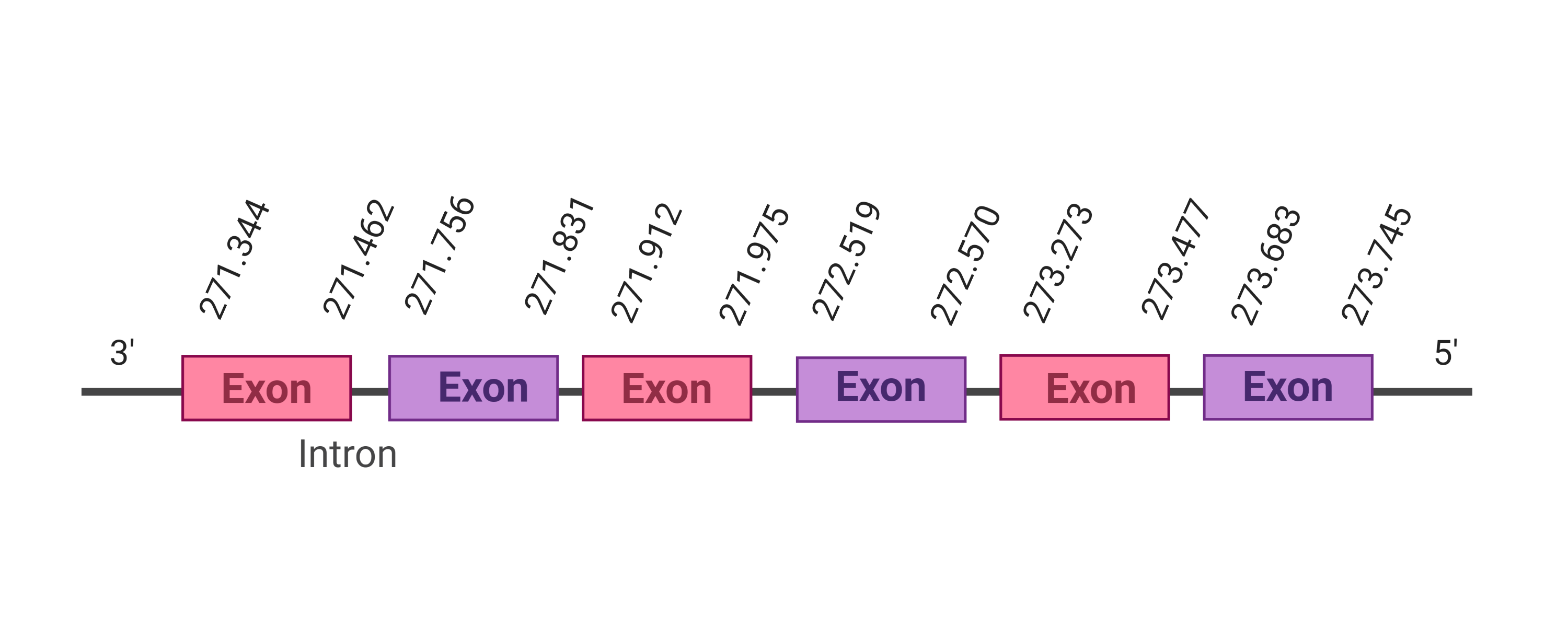
Thioredoxin reductase family
Thioredoxin reductases (TRs) control the redox state of thioredoxins. TRs are less dependent on dietary Se and are classified like housekeeping selenoproteins. Mammals have three TR isozymes, all of them are Sec-containing proteins [22]:
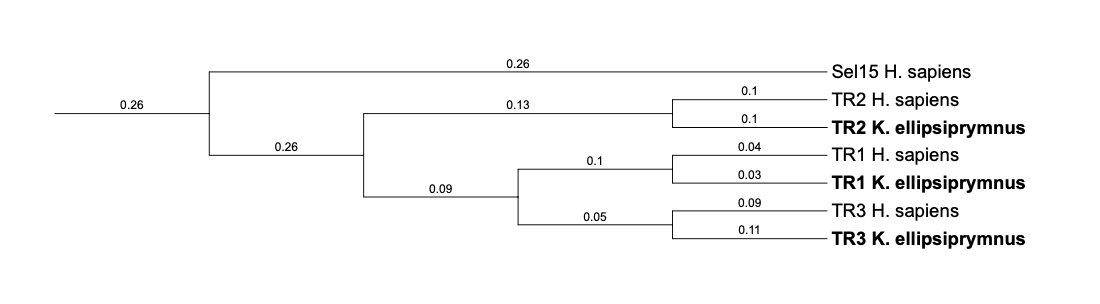
In the phylogenetic tree of Thioredoxin reductase family, it can be seen that the selenoproteins TR1, TR2 and TR3 predicted in K. ellipsiprymnus are classified together with their own queries. It suggests that the proteins have been predicted correctly.
TR1
TR2
TR3
eEFSec
In mammalians, the Sec-specific eukaryotic elongation factor (eFSec) is the elongation factor responsible for the delivery of the aminoacil-tRNASec into the ribosomal A site [26]. Interestingly, eEFSec binds to the SECIS-Binding protein 2 (SBP2), which binds to the SECIS element of the mRNAs. Both eEFSec and SBP2 are the proteins that are capable of inserting a Selenocysteine amino acid in a UGA codon [2].
SBP2
In mammalians, SECIS-Binding protein 2 is known to interact with the SECIS element, the ribosome and the eEFSec. Also, mutations in SBP2 are associated with abnormal thyroid function [2]. In some species, the amino-terminal domain of SBP2 is not present but they still contain selenocysteines in selenoproteins. Thus, it is thought that this part of the protein is not required for its function [33].
The protein TR1 is located in the scaffold SJYR01000959.1 (it was chosen the hit with the lowest e-value: 6.17e-40). The percentage of identity between the query and the hit is 95%. The protein is located in the positive strand between the nucleotide positions 701737 and 739796. In this protein, thirteen exons were predicted. When running tcoffee it was obtained a score of 1000 and visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that both the human protein SelW3 and the predicted protein in K. ellipsiprymnus conserve the Selenocysteine. The predicted protein starts with a methionine, which means that the beginning of the protein has been correctly predicted. Using Seblastian, the selenoprotein thioredoxin reductase 1 from Bos taurus was predicted in K. ellipsiprymnus. This protein has eight exons, and it is located in the positive strand. Also, one Secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein TR1 in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that TR1 is a selenoprotein.
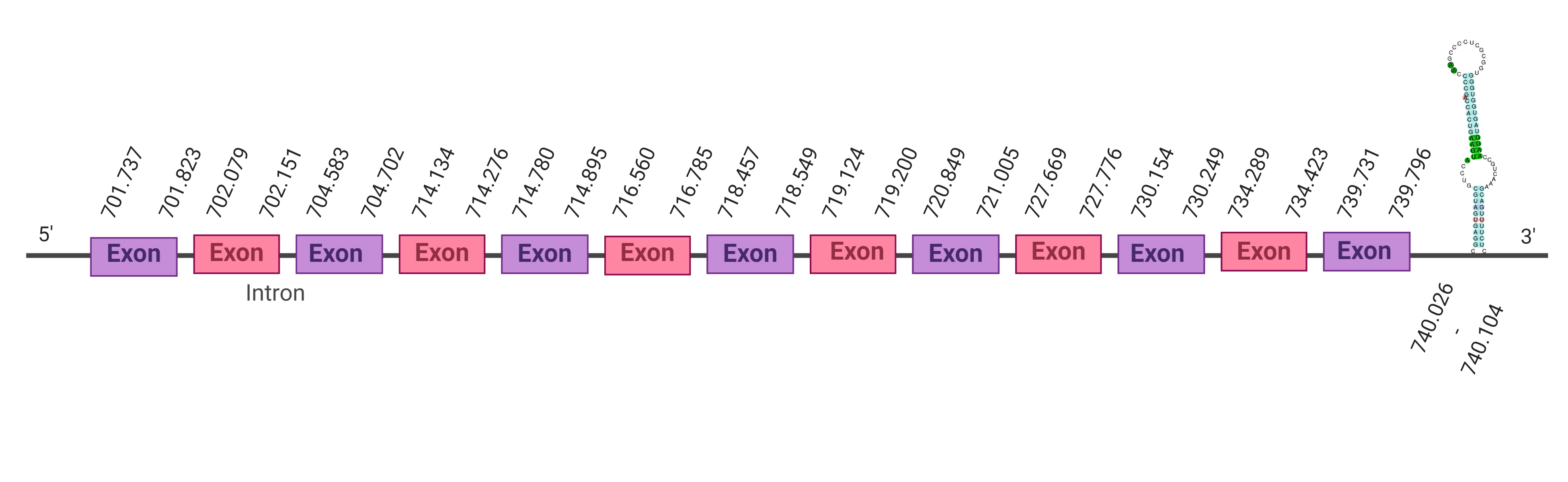
The protein TR2 is located in the scaffold SJYR01003863.1 (it was chosen the hit with the lowest e-value: 1.55e-22). The percentage of identity between the query and the hit is 95%. The protein is located in the negative strand between the nucleotide positions 69686 and 94531. In this protein, sixteen exons were predicted. When running tcoffee it was obtained a score of 983 and visualized a bad alignment with gaps inside the protein sequence. Other hits in the same scaffold were obtained, but all of them were analyzed when running the fastasubseq. The predicted TR2 protein contains a selenocysteine amino acid, but the beginning of the protein could not be correctly predicted (the predicted protein does not start with methionine). The human TR2 Selenoprotein also contains a selenocysteine.
As the predicted protein obtained from the human protein TR2 was not as good as it would have been expected, TR2 Selenoprotein was predicted again in K. ellipsiprymnus genome using the TR2 Bos taurus’ protein (Cow). Unfortunately, there’s no such protein for the Bos taurus. It didn’t appear neither in Uniprot nor in Selenodb.
No protein was predicted using the Seblastian, and no secis ele,ent was found. Taken all together, the protein TR2 in K. ellipsiprymnus (predicted from Homo sapiens’ TR2) contains selenocysteine but not a Secis element. All these signs would suggest that TR2 is not a selenoprotein.
The protein TR3 is located in the scaffold SJYR01001222.1 (it was chosen the hit with the lowest e-value: 1e-33). The percentage of identity between the query and the hit is 67%. The protein is located in the positive strand between the nucleotide positions 509806 and 565183. In this protein, seventeen exons were predicted. When running tcoffee, it was obtained a score of 982 and visualized a good alignment except at the beginning of the protein sequence, that couldn’t be found. Tcoffee aligns the human protein with the predicted protein and it could be observed that both the human protein TR3 and the predicted protein in K. ellipsiprymnus conserve the Selenocysteine. The predicted protein lacks a methionine at the beginning of the protein. Using Seblastian, the thioredoxin reductase 3 from Homo sapiens was predicted in K. ellipsiprymnus. This protein has eight exons, and it is located in the positive strand. Also, one Secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
Taken all together, the protein TR3 in K. ellipsiprymnus contains selenocysteine and a Secis element. All these signs would suggest that TR3 is a selenoprotein.
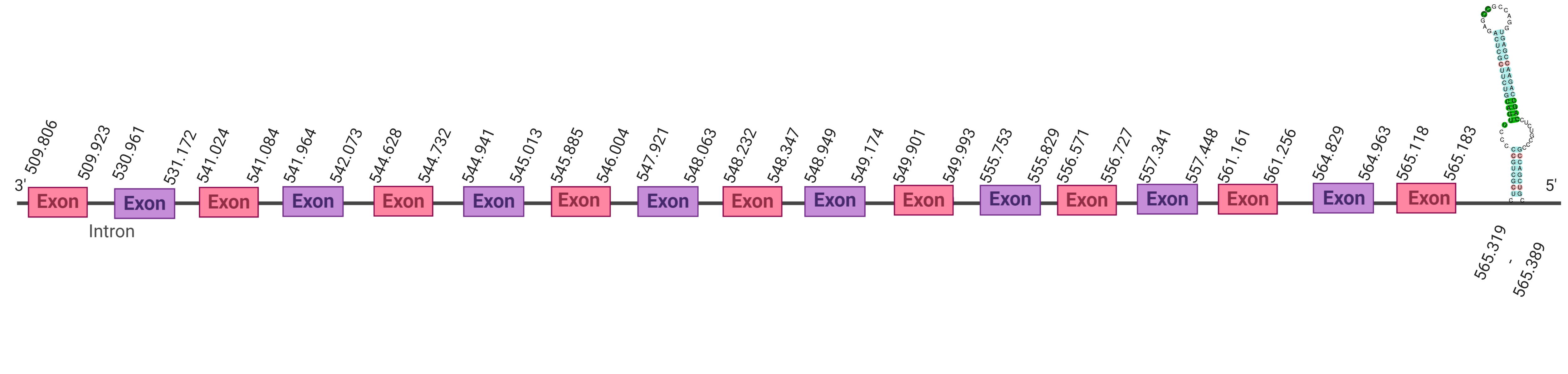
The protein eEFSec is located in the scaffold SJYR01000075.1 (it was chosen the hit with the lowest e-value: 6.18e-112). The protein is located in the negative strand between the nucleotide positions 1626365 and 1740483. In this protein, seven exons were predicted. When running tcoffee, it was obtained a score of 993 and visualized quite a good alignment. It has been correctly predicted all the protein, including the first methionine, but there are some mismatches between the predicted protein and the human eEFSec protein. It can be observed that neither the human protein eEFSec nor the predicted protein in K. ellipsiprymnus conserves the Selenocysteine. Any selenoprotein was predicted using the Seblastian, nor any Secis element was found in the predicted protein. This suggests that K. ellipsiprymnus’ eEFSec is not a selenoprotein.
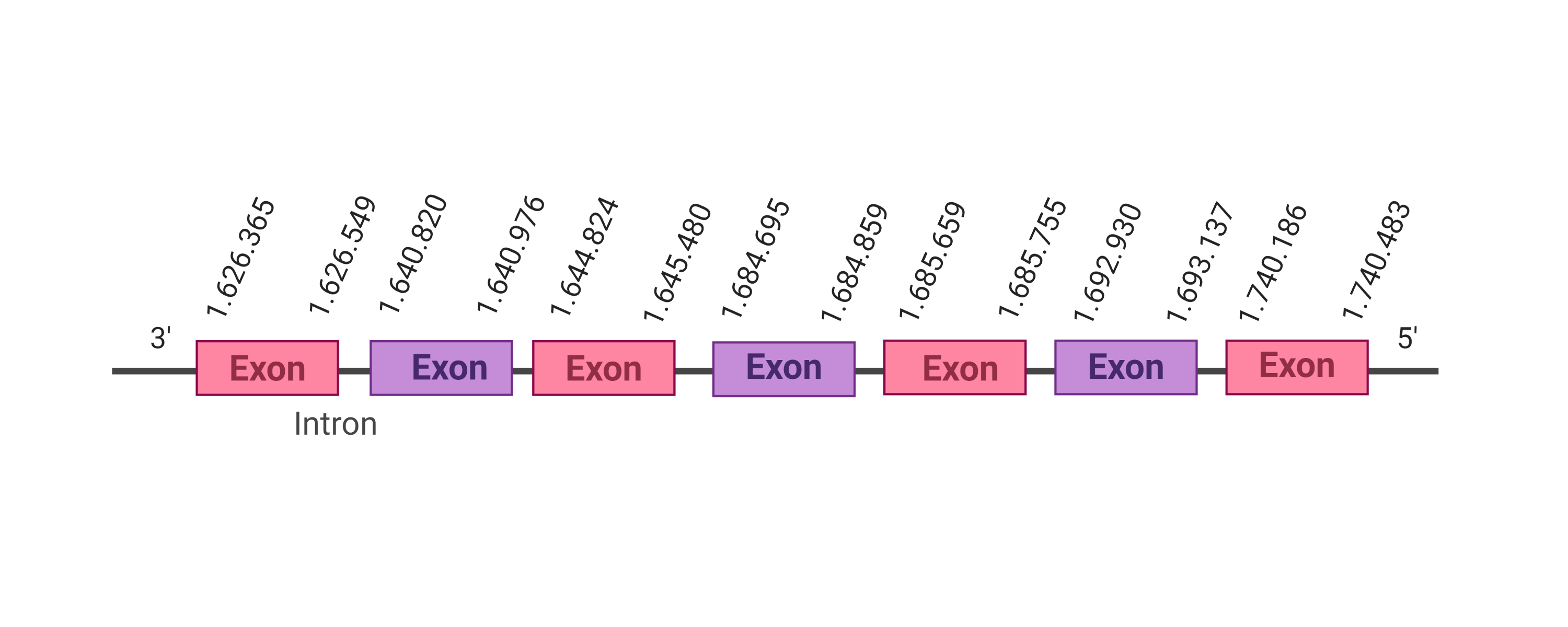
The protein SBP2 is located in the scaffold SJYR01003105.1 (it was chosen the hit with the lowest e-value: 2.04e-28). The percentage of identity between the query and the hit is 96%. The protein is located in the negative strand between the nucleotide positions 262147 and 287310. In this protein, sixteen exons were predicted. When running tcoffee, it was obtained a score of 993 and, visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that both the human protein SBP2 and the predicted SBP2 protein in K. ellipsiprymnus do not conserve the selenocysteine nor cysteine. The predicted protein lacks the first methionine amino acid at the beginning. Interestingly, as we have explained before, it has been reported that some species have been reported not to contain the amino-terminal domain of SBP2. K. ellipsiprymnus SBP2 protein seems to also have lost this region. No protein was predicted using the Seblastian. Additionally, any secis element was found in the negative strand.
Taken all together, the protein SBP2 in K. ellipsiprymnus does not contain a selenocysteine amino acid nor a Secis element. All these signs would suggest that nowadays SBP2 is not a selenoprotein.

Selenophosphate synthetase family
The selenophosphate synthetase family in mammals has the same function as the protein SelD from E. coli. Although both SPS1 and SPS2 are selenophosphatase synthetases, only SPS2 is capable of generating selenocysteines in vitro. Interestingly, SPS2 is thought to create de novo selenocysteines while SPS1 is thought to recycle selenocysteines [2].
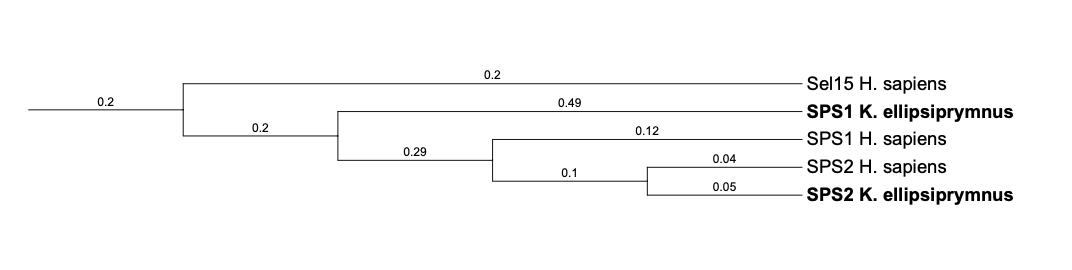
In the phylogenetic tree of Selenophosphate synthetase family, it can be seen that the predicted protein SPS2 in K. ellipsiprymnus is classified together with its own query. This suggests that SPS2 has been correctly predicted. However, the predicted protein SPS1 is not clustered with its query.
SPS1
SPS2
The protein SPS1 is located in the scaffold SJYR01001146.1 (it was chosen the hit with the lowest e-value: 0,0). The percentage of identity between the query and the hit is 72%. The protein is located in the positive strand between the nucleotide positions 26372 and 26266. In this protein, two exons were predicted. When running tcoffee, it was obtained a score of 995 and, visualized a perfect alignment. Tcoffee aligns the human protein with the predicted protein and it could be observed that the human protein SPS1 did not contain selenocysteine (it has a tyrosine instead) while the predicted SBP2 protein in K. ellipsiprymnus did conserve the selenocysteine. The predicted protein lacks the first methionine amino acid at the beginning of the protein, meaning that this region has not been correctly predicted. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein SPS1 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that SPS1 is a selenoprotein.
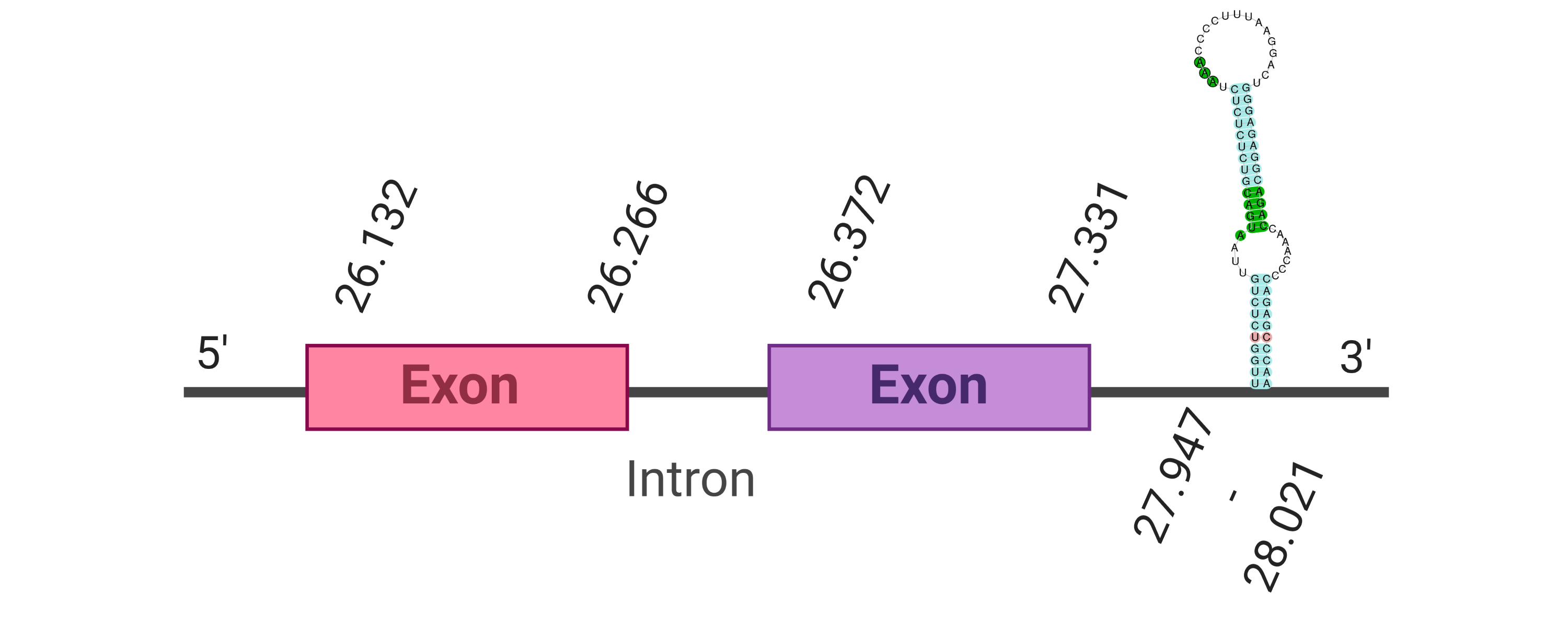
The protein SPS2 is located in the scaffold SJYR01001146.1 (it was chosen the hit with the lowest e-value: 0,0). The percentage of identity between the query and the hit is 92%. The protein is located in the positive strand between the nucleotide positions 26114 and 27352. In this protein, one exon was predicted. When running tcoffee, it was obtained a score of 998 and, visualized a perfect alignment. The whole selenoprotein was correctly predicted. The predicted protein starts with a methionine.Tcoffee aligns the human protein with the predicted protein and it can be observed that both of them conserve the Selenocysteine amino acid. No protein was predicted using the Seblastian, but one secis element (grade A) in the positive strand was correctly predicted in the 3’ sequence of the protein.
The protein SPS2 in K. ellipsiprymnus contains a selenocysteine and a Secis element. All these signs would suggest that SPS2 is a selenoprotein.
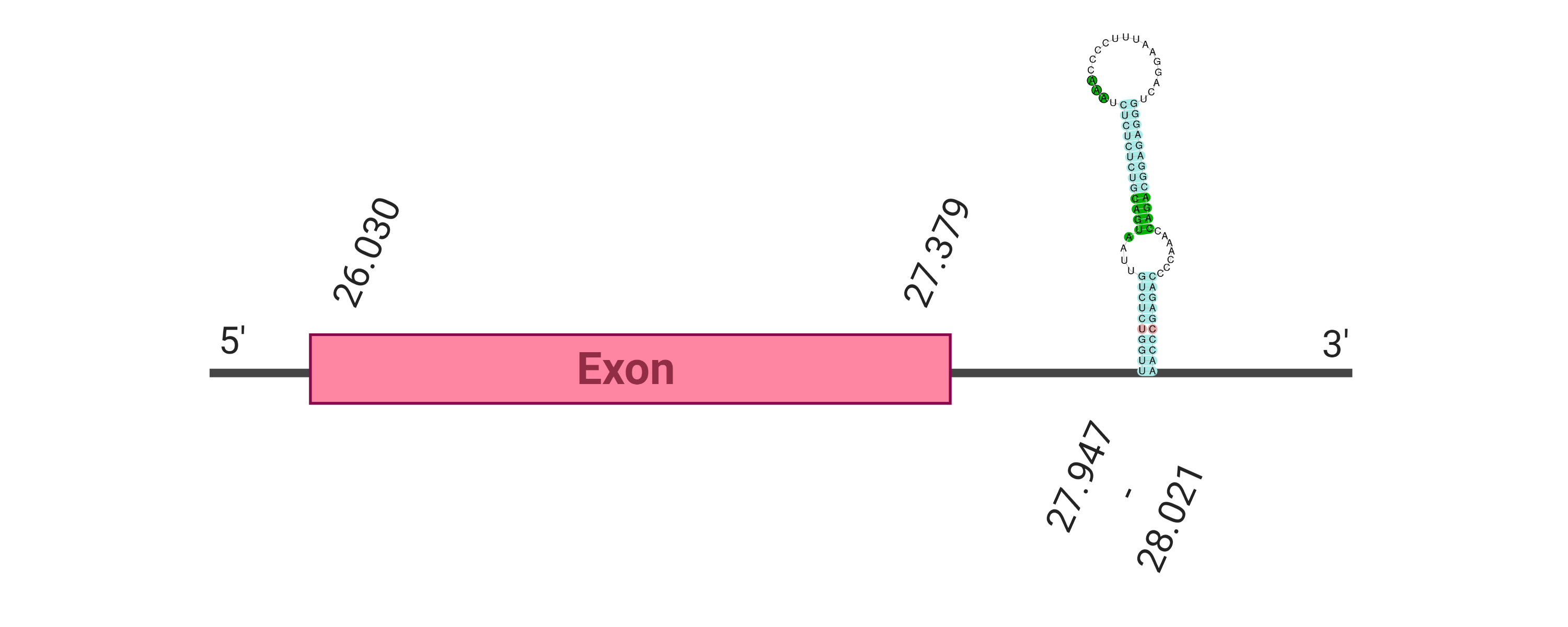
The purpose of this paper has been to contribute to the annotation of the Kobus ellipsiprymnus genome, by annotating the Selenoproteins present in its genome. In order to address such aim, human Selenoproteins homologous were searched in Kobus ellipsiprymnus genome. The reason why the human genome was chosen as the genome of reference is that it is one of the best annotated genomes. If chosen a genome of reference closer phylogenetically to our animal, it wouldn’t be so well annotated, and therefore many existing selenoproteins wouldn’t have been checked.
A program was written in order to do so. Such program would ask for a human Selenoprotein (the query) and would look for its homology on the Kobus ellipsiprymnus genome, suggesting different hits, therefore suggesting different options of the prediction of the protein. One of the hits will be selected and the program will run tcoffee, which aligns the predicted protein with the human Selenoprotein (the query), and prints both the alignment and its corresponding score. All this information allows us to consider if it exists an homologous of the given human Selenoprotein on the Kobus ellipsiprymnus genome, and, if so, if it is conserved (it contains Selenocysteine amino acid) or it has been lost. Additionally, a Selenoprotein Prediction Server was used in order to address whether there were secis elements in the predicted proteins. Also, this tool checked if there was any selenoprotein in the given sequence that match with a selenoprotein database.
From a total of 38 selenoproteins checked, the characterization is the following one:
*Concerning SelN. The human selenoprotein contained two Selenocysteines while the predicted protein only one (the other one, instead, turns out to be an Alanine).
From those predicted selenoproteins, some of them have been predicted including the first methionine amino acid and others not:
| Selenoproteins with Met | Selenoproteins without Met |
|---|---|
| GPx2, GPx3, GPx4, DI1, DI2, DI3, SPS2, SelH, SelI, SelK, SelM, SelP, SelR1, SelT, TR1, TR2. | GPx1, GPx6, SPS1, Sel15, SelN, TR3. |
Methionine was not correctly predicted at the beginning of each protein for different reasons. By doing an homology-based approach, it was assumed that the protein present in K. ellipsiprymnus starts at the same point as the human protein. Also, the identification of another amino acid where it was supposed to be methionine may be explained due to the presence of misanotations in the K. ellipsiprymnus’ genome.
With the program and methodology explained above, 29 of the 38 proteins evaluated were identified on the K. ellipsiprymnus genome: 22 selenoproteins, 7 cysteine-homologous proteins and 4 selenoprotein machinery proteins were identified. Among these proteins, some particularities were found:
Optimization of the code
Once the program was written down, and knowing that it would have to be run at least once for each existing human selenoprotein we tried to iterate it as much as possible. All the commands were enclosed in an loop. At the same time, a vector was created. Each human selenoprotein was saved in a different position of the vector. The loop was repeated as many times as positions had the vector, meaning that every single selenoprotein was evaluated, and homologous on the Kobus ellipsiprymnus genome were looked for each of them.
Another optimization of the code was to teach the program to calculate on his own the length in which it was enclosed the hit, information that was required later on when calling the program fastasubseq. Knowing both the starting point of the hit and the length of the scaffold in which it was embedded, the program would make sure that the value that takes length every time does not exceed the size of the particular scaffold.
However, due to the shortage of time and our limited knowledge there were many things that we didn’t reach to automatize, even though we wanted and tried. We weren’t capable of telling the program to read all the Selenoprotein files contained in one folder while saving them on different vector’s positions, and instead, we had to introduce manually the name of the all Selenoproteins that had to contain the vector.
Limitations of the followed method
Given that the annotation of Kobus ellipsiprymnus selenoproteins in based on an homologous human selenoproteins search, we will encounter some limitations. The first one is the fact that Selenoproteins not present in humans will not be taken into consideration. If there is any Selenoprotein present in the genome of Kobus ellipsiprymnus but it doesn’t appear on the human genome, it won’t be looked for in this study.
Moreover, not all hits that tblastn had given us for all the selenoproteins checked have been evaluated, again, due to the shortage of time. Only those of them that had low e-values and high percentage of identities have been evaluated. However, some duplications or triplications, or partial duplications or triplications might have been disregarded because of this.
Additionally, the exons of each protein were predicted with exonerate, which looks for all the exons present in a given sequence (extracted from fastasubseq). Therefore, if the given sequence is bigger than the protein itself that we are looking for, more exons will be predicted (exons that no not belong to the protein).
The tool Seblastian was used to check the presence of secis elements in the predicted proteins. Seblastian also looks for known selenoproteins in the given sequence, however it only predicted as a selenoprotein 11 of 38 proteins (and not all of them correctly). Therefore, the results obtained in this study suggest that the optimal method to characterize selenoproteins is by an homology-approach.
There are many people that need to be thanked for making this project possible. First of all our tutor Aitor Serres who has been there always solving our doubts and problems. Without him, the program optimization would have been much weaker.
Secondly, thanks to all the tutors' team, as all of them have supported and guided us through all the way.
Thirdly, we want to thank Romain Gudimard for showing interest on the project.
Finally, thanks to the classmates for having helped each other, the good work environment during the long afternoons, and the Christmas songs played during the work sessions.
Universitat Pompeu Fabra Campus del Mar
Carrer del Dr. Aiguader, 80
08003 Barcelona, Spain