RESULTS
In the following table you can see all our results:
Photo |
Info |
||||||||||
| Iodothyronine deiodinases | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
1699 - 2819 (+) |
|
|
|
|
|||
| |
|
|
|
819 - 959 (-) |
|
|
|
|
|||
| GPX Family | |||||||||||
| |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
|
|
|
|
|
|
||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|||||||
| Fep15 | |||||||||||
| |
|
|
|
|
|
|
|
||||
| MsrA | |||||||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| Sel | |||||||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
|
|||||
| TR | |||||||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|
||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
||||||
Photo |
Info |
||||||||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| (trnau1ap*) |
|
|
|
|
|
|
|
|
|||
(trau1ap-like) |
|
|
|
|
|
|
|
||||
| |
|
|
|
|
|||||||
(trnau1ap-like~) |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
|
|||
| |
|
|
|
|
|
|
|||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|||||||
| |
|
|
|
|
|
|
|
|
Result analysis of selenoproteins
and machinery
and machinery
Below, it is provided the structural data about the selenoprotein’s genes found in tuna, as well as schematic drawing, where it is shown the following information:
- Genes drawing from the 5’ to the 3’ side
- Number of exons of each gene
- Number of the first and the last nucleotide of each exon
- Contigs where each gene has been predicted (shown in different colors)
- Gene exon where the Sec is found
- SECIS element/s found in each selenoprotein represented
It can not be assured that the schematic information provided is completely reliable, as not all selenoproteins have been entirely predicted. Nevertheless, they do depict the data obtained by means of our methodology.
Iodothyronine deiodinases (DI)
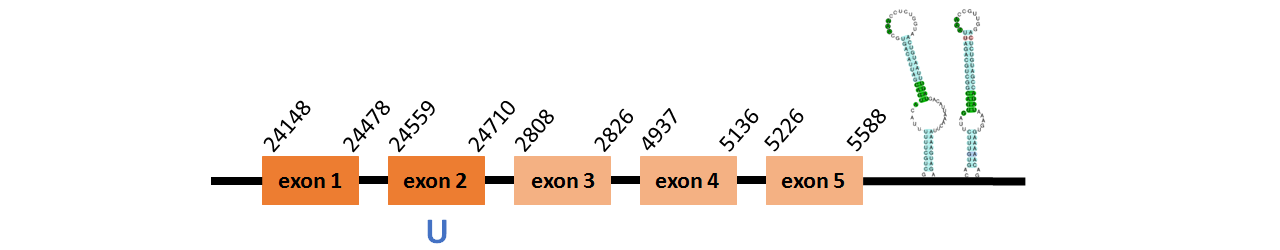
Protein DI1 is located in contig number BADN01054393.1 between positions 24148 and 24710 and in contig number BADN01054394.1 between positions 2808 and 5588, both of them in the forward strand. The gene contains 5 exons, with a Selenocystein in exon 2.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish DI1 protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found 2 SECIS elements, both of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
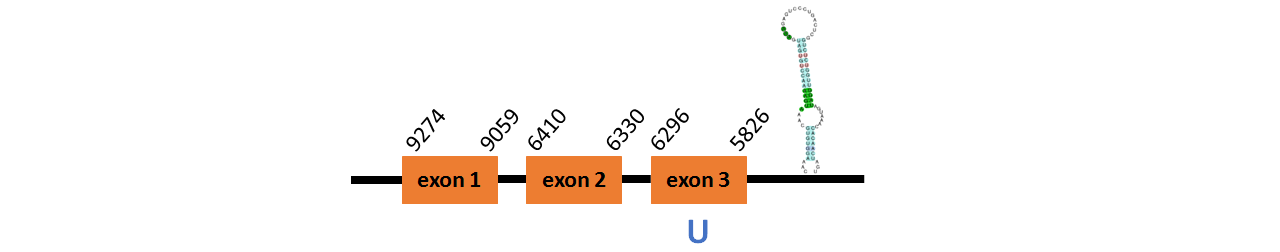
Protein DI2 is located in contig number BADN01070230.1 between positions 5826 and 9274 in the reverse strand. The gene contains 3 exons, with a Selenocystein in exon 3.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish DI protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
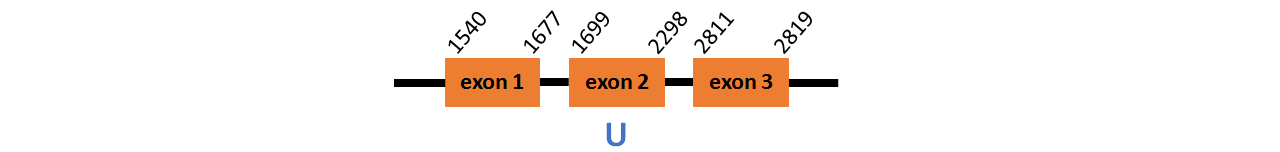
Protein DI3a is located in contig number BADN01059959.1 between positions 1540 - 1677 and
1699 - 2819 in the forward strand. The gene contains 3 exons, with a Selenocystein in exon 2.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the human DI protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS element in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein DI3b is located in contig number BADN01080856.1 between positions 219 - 824 and
819 - 959 in the reverse strand. The gene contains 2 exons, with a Selenocystein in exon 2.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the human DI protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
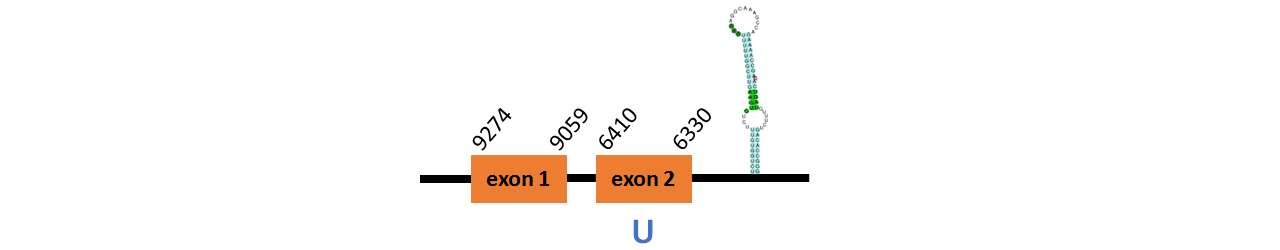
Protein GPx1a is located in contig number BADN01004052.1 between positions 3443 and 4841 in the forward strand. The gene contains 2 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
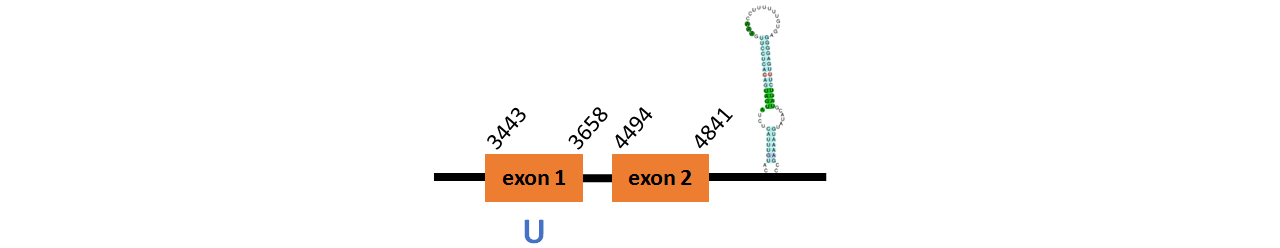
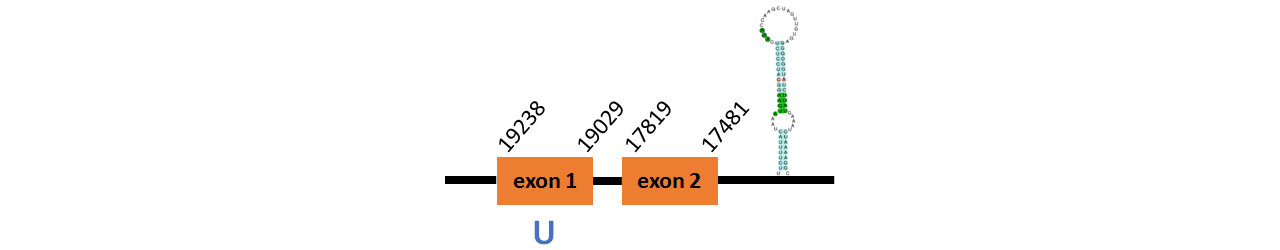
Protein GPx1b is located in contig number BADN01015002.1 between positions 17481 and 19238 in the reverse strand. The gene contains 2 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
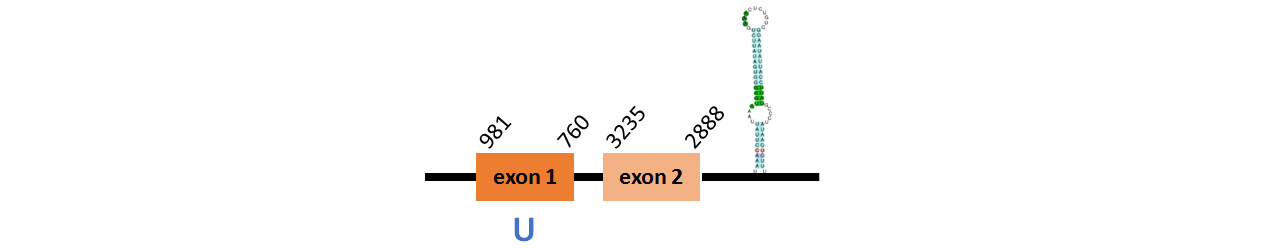
Protein GPx2 is located in contig number BADN01015930.1 between positions 2888 and 3235 in the reverse strand and in contig number BADN01015931.1 between positions 760 and 981 in the reverse strand. The gene contains 2 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish DI2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
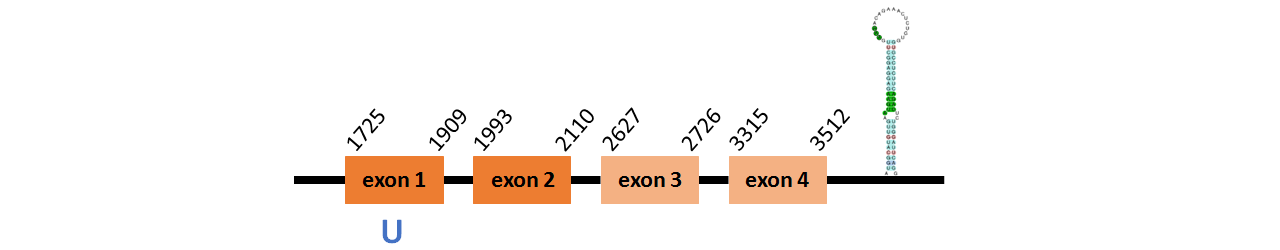
Protein GPx3 is located in contig number BADN01020040.1 between positions 1725 and 3512 in the forward strand. The gene contains 4 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx3 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
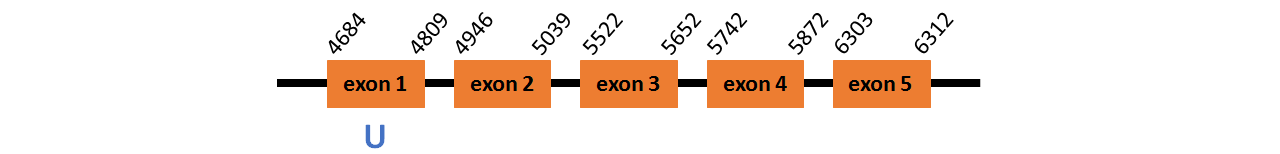
Protein GPx3b is located in contig number BADN01011894.1 between positions 4684 and 6312 in the forward strand. The gene contains 5 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
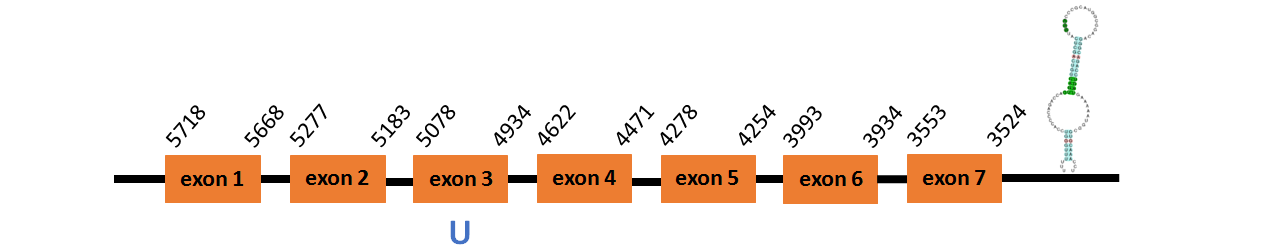
Protein GPx4a is located in contig number BADN01049899.1 between positions 3524 and 5718 in the reverse strand. The gene contains 7 exons, with a Selenocystein in exon 3.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx4 protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
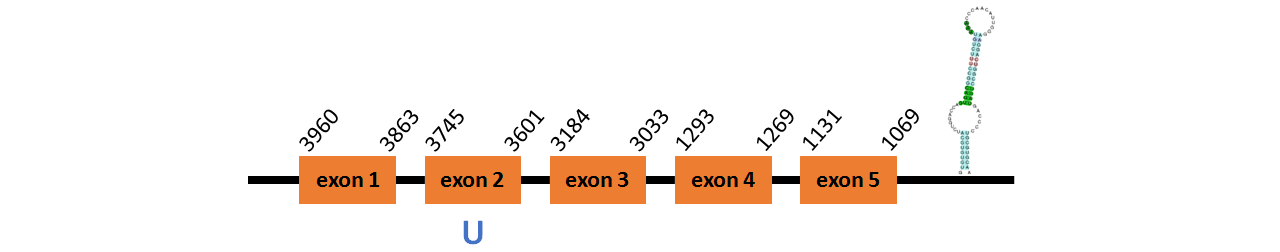
Protein GPx4b is located in contig number BADN01052654.1 between positions 1069 and 3960 in the reverse strand. The gene contains 5 exons, with a Selenocystein in exon 2.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein GPx7 is located in contig number BADN01103388.1 between positions 4320 and 4391 in the forward strand. The gene contains 3 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human GPx7 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

Protein GPx8 is located in contig number BADN01071155.1 between positions 1526 and 2166 and in contig number BADN01071156.1 between positions 29 and 199, both of them in the forward strand. The gene contains 3 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish GPx8 protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

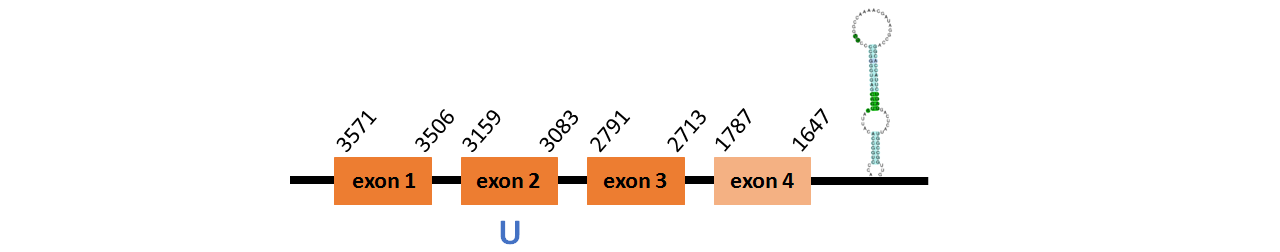
Protein Fep15 is located in contig number BADN01014294.1 between positions 1647 and 3571 in the reverse strand. The gene contains 4 exons, with a Selenocystein in exon 2.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish Fep15 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein MsrA1 is located in contig number BADN01102191.1 between positions 305 and 451, in contig number BADN01114924.1 between positions 738 and 1271, in contig number BADN01114925.1 between positions 1979 and 3410, all of them in the forward strand, and in contig number BADN01116844.1 between positions 406 and 6998, in the reverse strand. The gene contains 7 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish MsrA protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
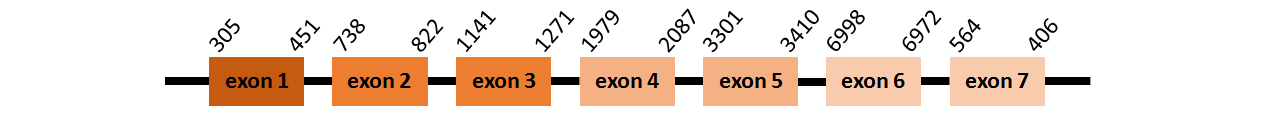
Protein MsrA2 is located in contig number BADN01022495.1 between positions 2007 and 2129, in contig number BADN01022497.1 between positions 845 and 2474 and in contig number BADN01022498.1 between positions 1867 and 2197, all of them in the reverse strand. The gene contains 5 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish MsrA protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

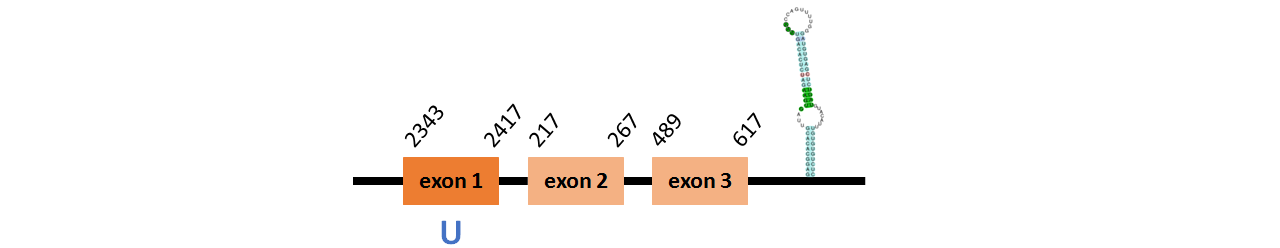
Protein Sel15 is located in contig number BADN01010583.1 between positions 2343 and 2417 and in contig number BADN01010584.1 between positions 217 and 617, both of them in the forward strand. The gene contains 3 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish DI1 protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of them of grade A, predicted in the 3’UTR region.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
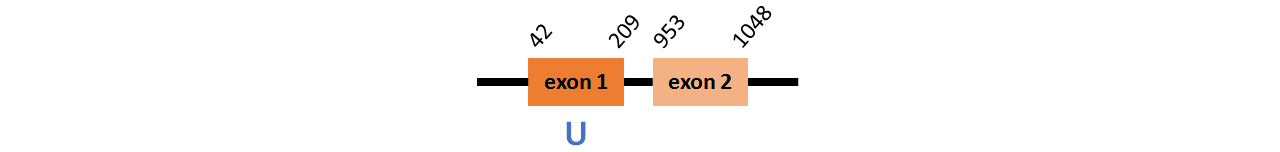
Protein SelH is located in contig number BADN01109579.1 between positions 42 and 209 and in contig number BADN01109580.1 between positions 953 and 1048, both of them in the forward strand. The gene contains 2 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelH protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
Protein SelH~ is located in contig number BADN01109583.1 between positions 42 and 1519 in the forward strand. The gene contains 2 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelH protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
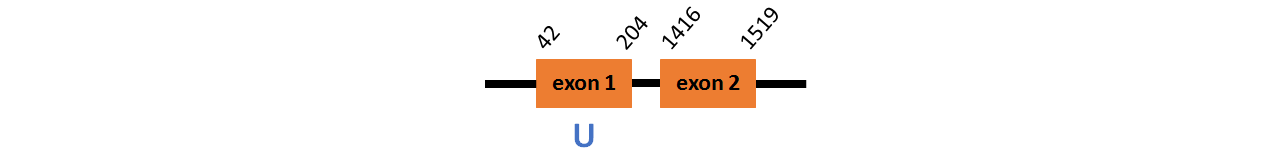
Protein SelI (cept1a~) is located in contig number BADN01047755.1 between positions 3 and 7887 in the reverse strand. The gene contains 9 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human SelI protein SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
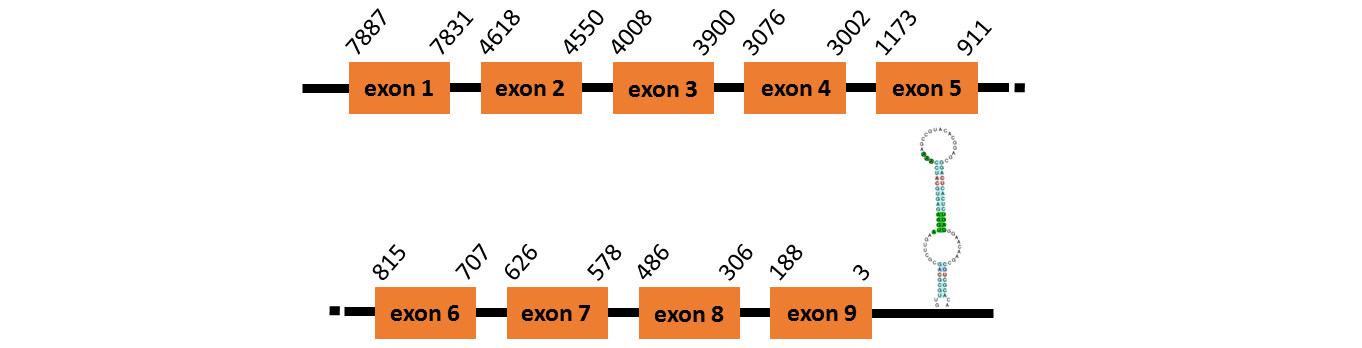
Protein SelJ is located in contig number BADN01033484.1 between positions 3077 and 4744 and in contig number BADN01033485.1 between positions 152 and 1231, both of them in the forward strand . The gene contains 9 exons, with a Selenocystein in exon 7.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelJ protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
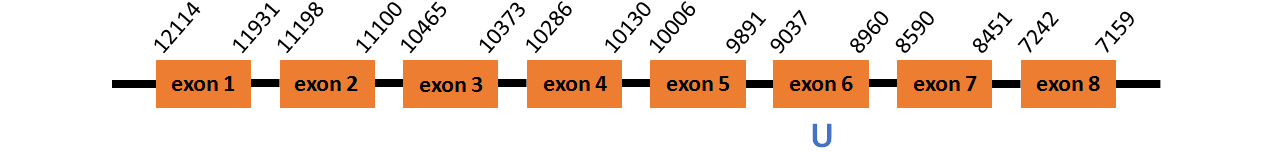
Protein SelJ~ is located in contig number BADN01079312.1 between positions 7159 and 12114 in the reverse strand . The gene contains 8 exons, with a Selenocystein in exon 6.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelJ protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
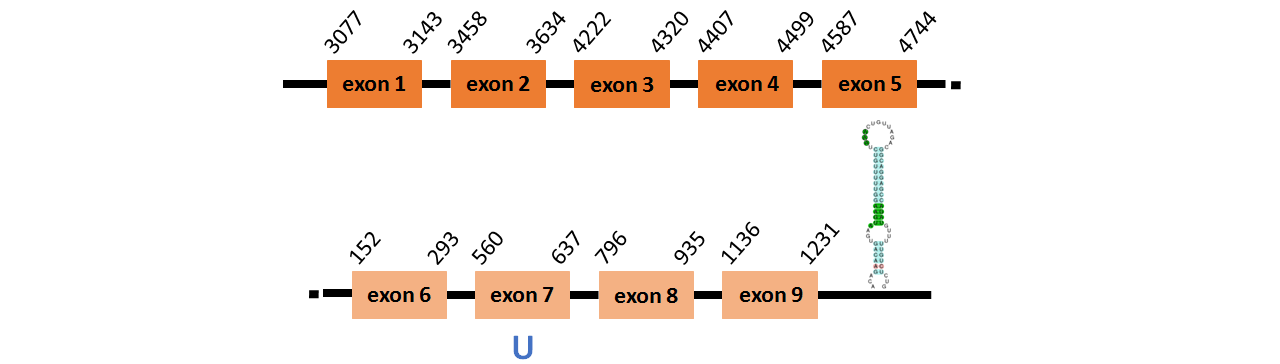
Protein SelK is located in contig number BADN01083792.1 between positions 3077 and 4744 and in contig number BADN01033485.1 between positions 152 and 1231, both of them in the forward strand . The gene contains 9 exons, with a Selenocystein in exon 7.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelK protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
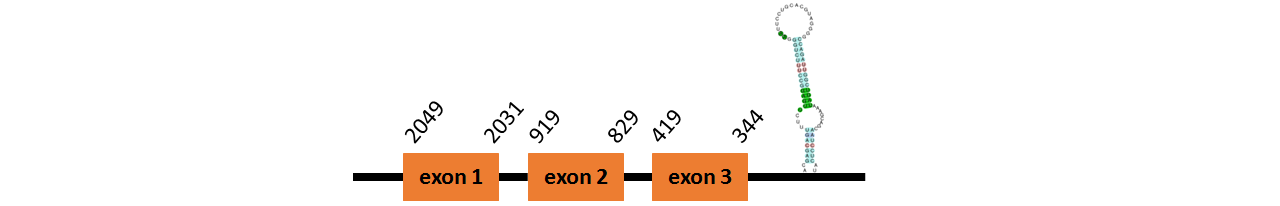
Protein SelL is located in contig number BADN01093036.1 between positions 1592 and 1814, in contig number BADN01093037.1 between positions 786 - 968 and in contig number BADN01093038.1 between positions 77 - 181, all of them in the forward strand. The gene contains 4 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelL protein found in SelenoDB.
We found 2 SECIS elements, one of them grade A and the other one grade C, predicted in the 3’UTR region.
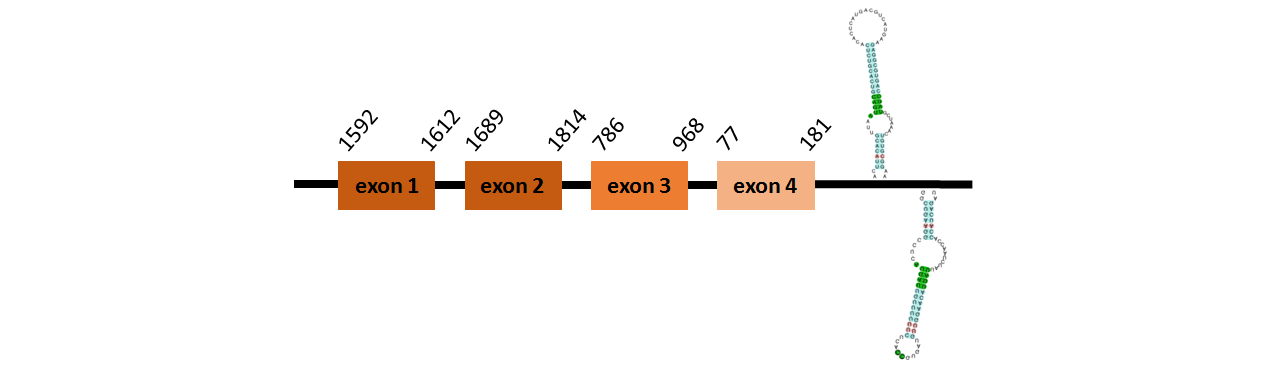
Protein SelL~ is located in contig number BADN01079553.1 between positions 2197 and 8391 in the forward strand. The gene contains 8 exons, but does not include any Selenocysteins.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelL protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

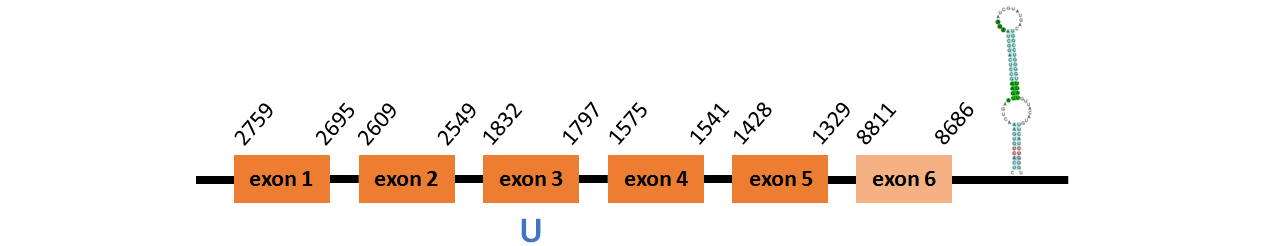
Protein SelM is located in contig number BADN01035956.1 between positions 1592 and 1814 in the forward strand, and in contig number BADN01035957.1 between positions 1329 and 2759 in the reverse strand. The gene contains 6 exons, with a Selenocystein in exon 3.
This protein was predicted blasting Thunnus orientalis genome against the human SelM protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
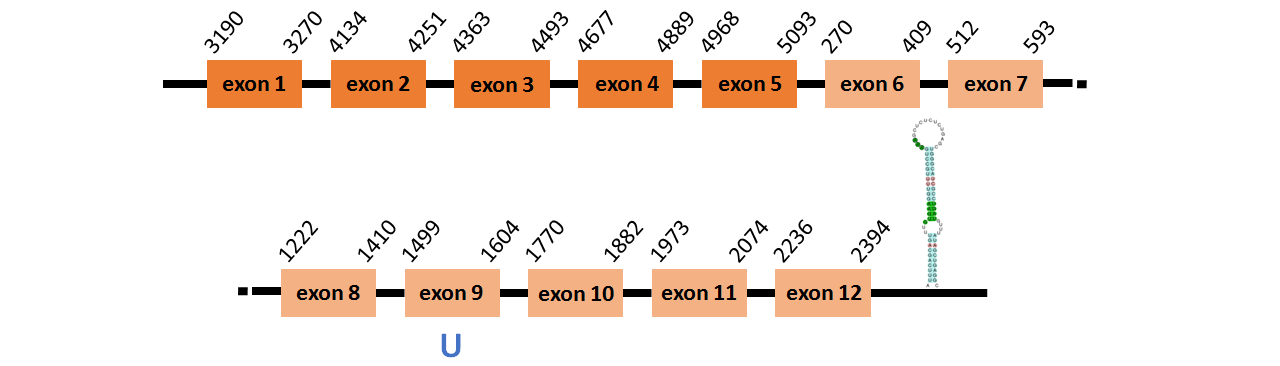
Protein SelN is located in contig number BADN01094294.1 between positions 3190 and 5096 and in contig number BADN01094295.1 between positions 270 and 2394, both of them in the forward o strand. The gene contains 12 exons, with a Selenocystein in exon 9.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelN protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein SelO is located in contig number BADN01010693.1 between positions 1155 and 2229 and in contig number BADN01010694.1 between positions 537 and 3814, both of them in the reverse strand. The gene contains 9 exons, with a Selenocystein in exon 9.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelO protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
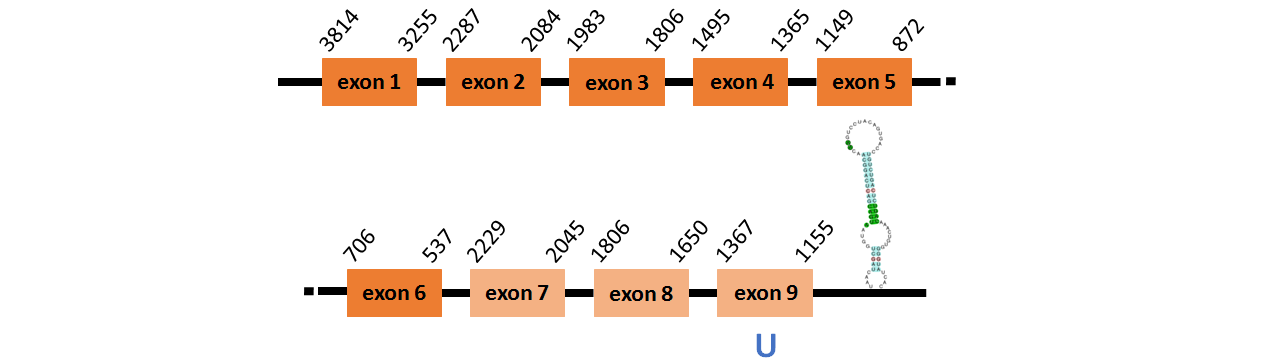
Protein SelO~ is located in contig number BADN01079895.1 between positions 403 and 2275 and in contig number BADN01079896.1 between positions 29 and 13896, both of them in the forward strand. The gene contains 13 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelO protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
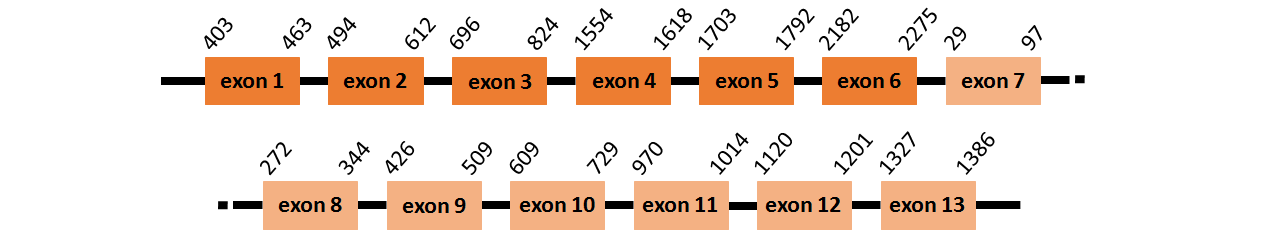
Protein SelO2 is located in contig number BADN01020657.1 between positions 46 and 5768 in the forward strand. The gene contains 9 exons but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelO protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found two SECIS elements, one of them of grade A and the other of grade B, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
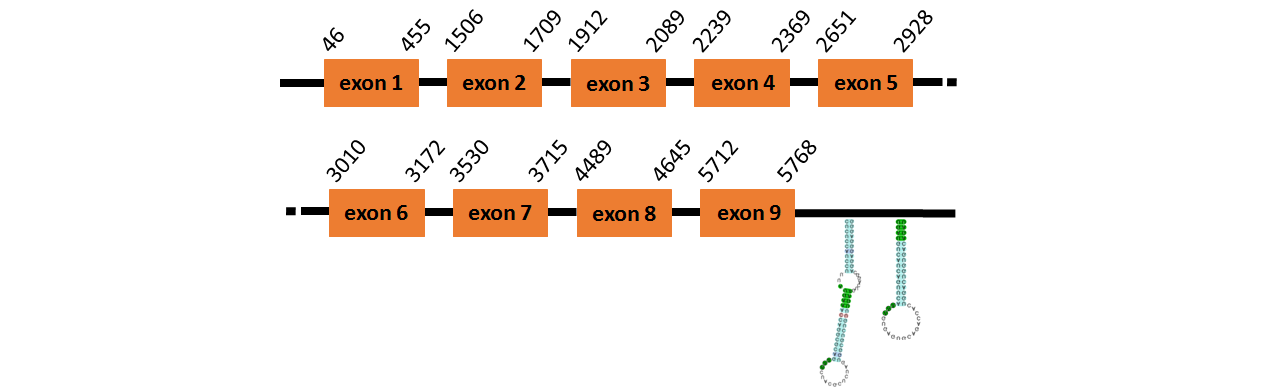
Protein SelP1a is located in contig number BADN01026358.1 between positions 6819 and 8589 in the reverse strand. The gene contains 5 exons, with a Selenocystein in exon 1 and 12 Selenocysteins in exon 5.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelP protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found two SECIS elements, one of them of grade A and the other of grade B, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
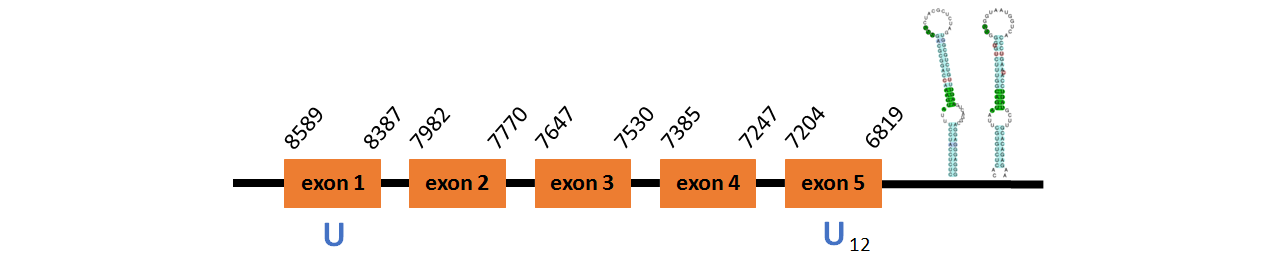
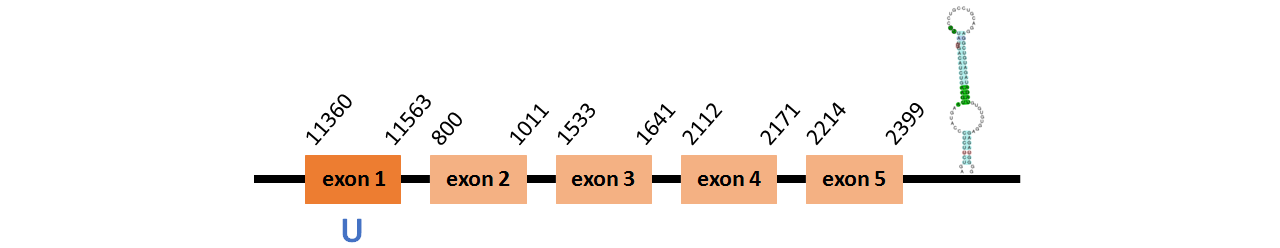
Protein SelP1b is located in contig number BADN01078432.1 between positions 11360 and 11563 and in contig number BADN01078433.1 between positions 800 and 2399, both of them in the forward strand. The gene contains 5 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelP protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
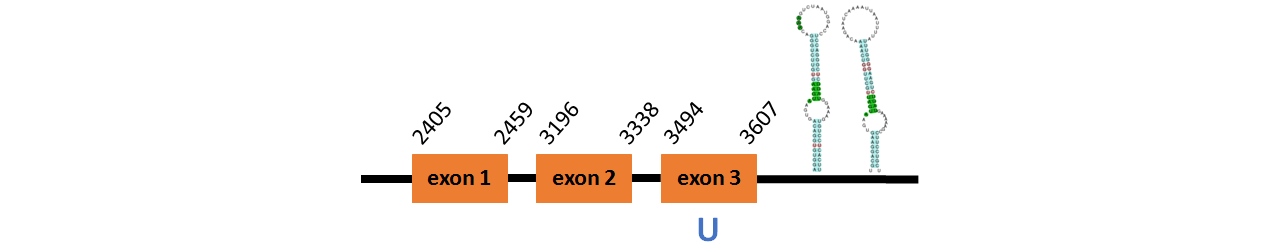
Protein SelR1a is located in contig number BADN01013935.1 between positions 2405 and 3607 in the forward strand. The gene contains 3 exons, with a Selenocystein in exon 3.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelR protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found two SECIS elements, one of them of grade B and the other of grade C, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
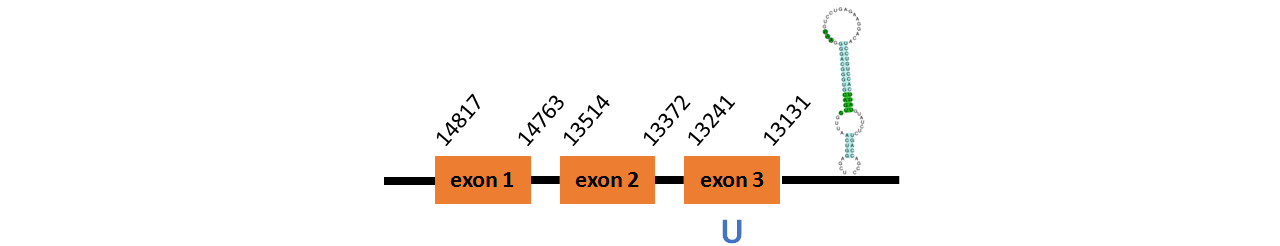
Protein SelR1b is located in contig number BADN01040629.1 between positions 13131 and 14817 in the reverse strand. The gene contains 3 exons, with a Selenocystein in exon 3.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelR protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A, predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
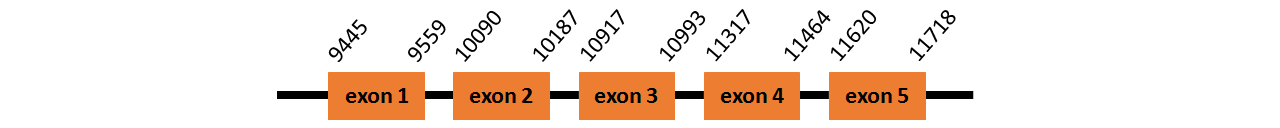
Protein SelR2 is located in contig number BADN01055509.1 between positions 9445 and 11718 in the forward strand. The gene contains 5 exons but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelR2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein SelR3 is located in contig number BADN01030477.1 between positions 6471 and 13602 and in contig BADN01030478.1 between positions 2433 and 2510, both of them in the reverse strand. The gene contains 6 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelR3 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

Protein SelS is located in contig number BADN01048450.1 between positions 422 and 4521 in the reverse strand. The gene contains 5 exons but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.

Protein SelT1a is located in contig number BADN01025729.1 between positions 6197 and 6628 and in contig number BADN01025730.1 between positions 1010 and 5399, both of them in the reverse strand. The gene contains 5 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelT protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
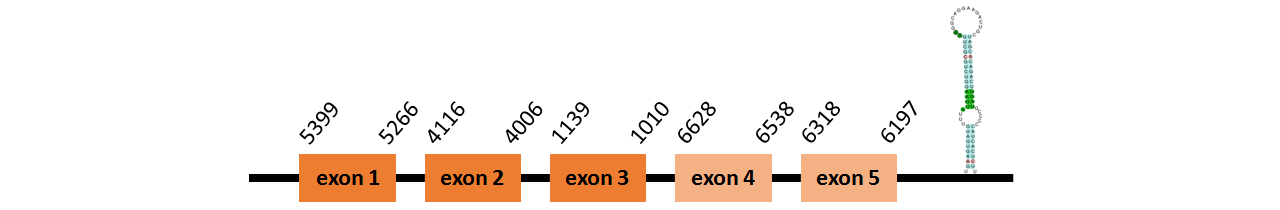
Protein SelT1b could not be predicted in Thunnus orientalis genome using our methodology or Selenoprofiles, even though it is found in the zebrafish proteome.
Protein SelT2 is located in contig number BADN01112525.1 between positions 1992 and 2568 and in contig number BADN01112526.1 between positions 30 and 1390, both of them in the forward strand. The gene contains 6 exons, with a Selenocystein in exon 3.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelT protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
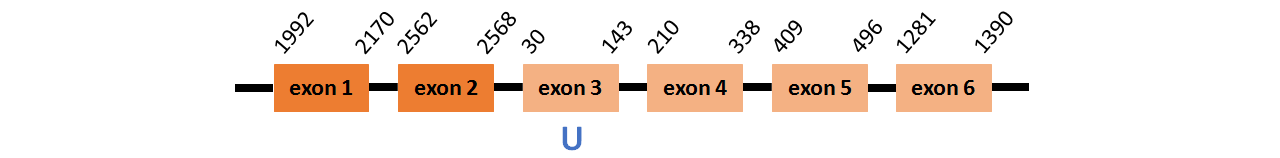
Protein SelU1a is located in contig number BADN01036881.1 between positions 2270 and 3201 and in contig number BADN01036882.1 between positions 1128 and 1220, both of them in the reverse strand. The gene contains 5 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the human SelU protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS elements of grade A predicted in the 3’UTR region.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
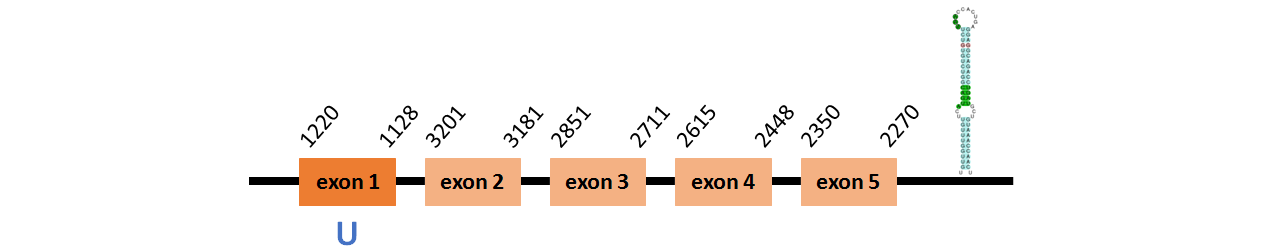
Protein SelU1b is located in contig number BADN01076486.1 between positions 2018 and 4049 in the forward strand. The gene contains 5 exons, with a Selenocystein in exon 2.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the human SelU protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
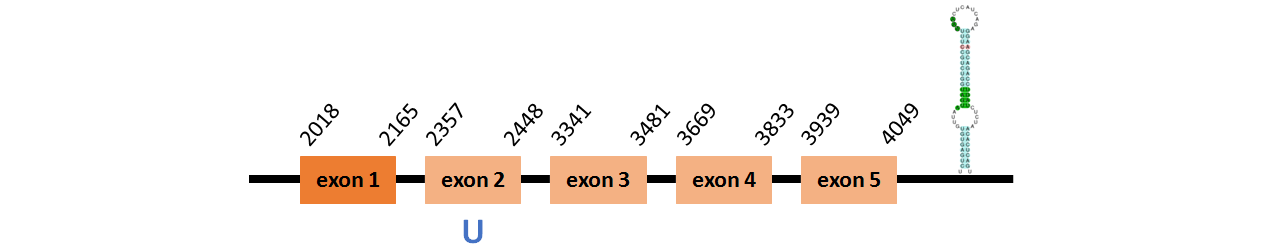
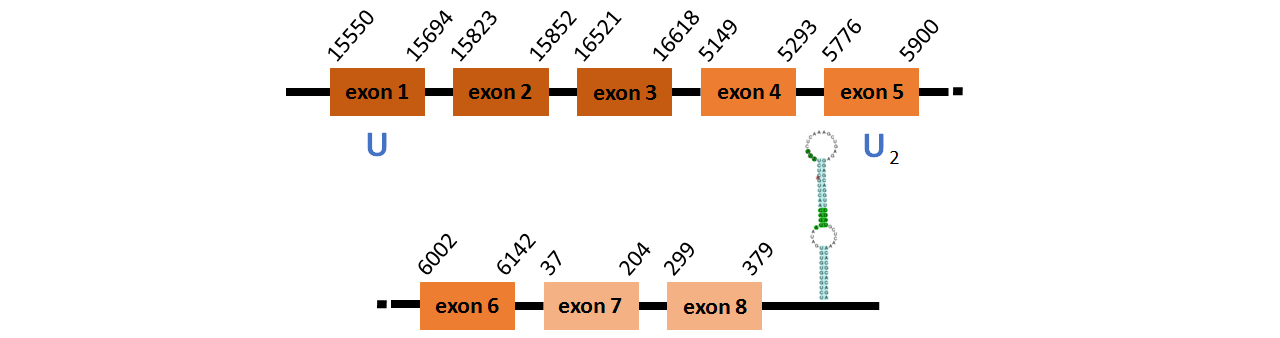
Protein SelU1c is located in contig number BADN01045672.1 between positions 15550 and 16618, in conting number BADN01045673.1 between positions 5149 and 6142, and in conting BADN01045674.1 between positions 37 and 379, all of them in the forward strand. The gene contains 8 exons, with a Selenocystein in exon 1.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelU protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein SelU2 is located in contig number BADN01089330.1 between positions 542 and 1254 in the forward strand. The gene contains 4 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human SelU2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.

Protein SelU3 is located in contig number BADN01042822.1 between positions 164 and 841 in the reverse strand. The gene contains 2 exons but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human U3 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
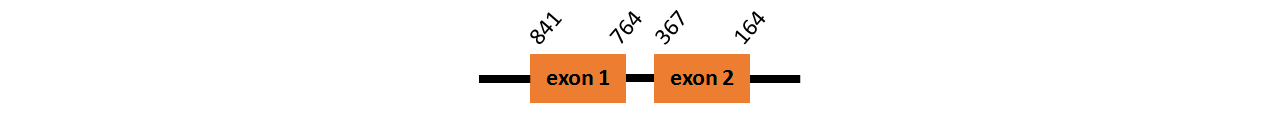
Protein SelW1 could not be predicted in Thunnus orientalis genome using our methodology or Selenoprofiles, even though it is found in the zebrafish proteome.
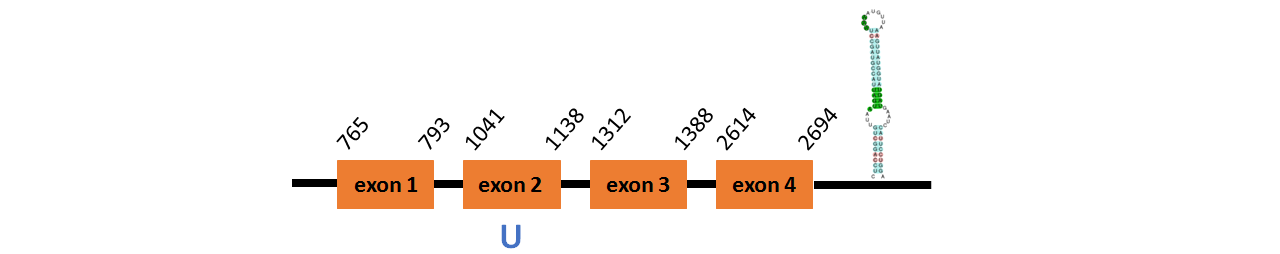
Protein SelW2a is located in contig number BADN01032869.1 between positions 765 and 2694 in the forward strand. The gene contains 4 exons, with a Selenocystein in exon 2.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SelW protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein SelW2b could not be predicted in Thunnus orientalis genome using our methodology or Selenoprofiles, even though it is found in the zebrafish proteome.
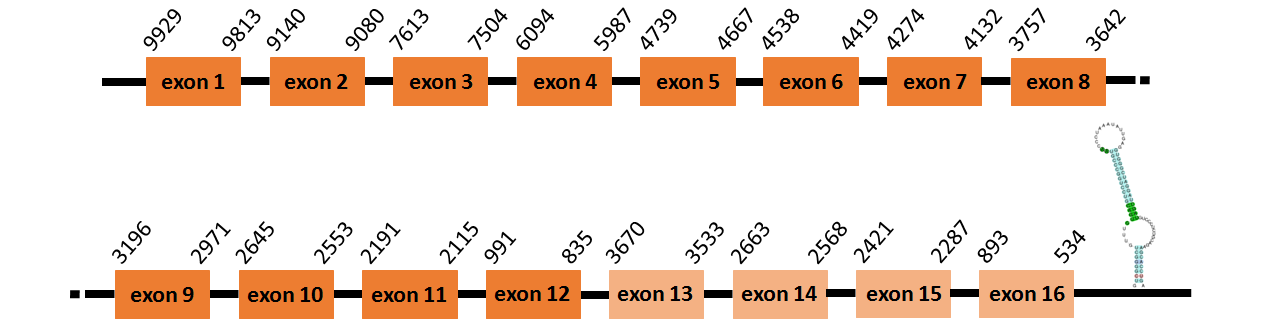
Protein TR1 is located in contig number BADN01025333.1 between positions 834 and 3670 and in contig number BADN01025334.1 between positions 835 and 9929, both of them in the reverse strand. The gene contains 16 exons but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish TR1 protein found in UniProt, because we could not find any information about it in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein TR1~A is located in contig number BADN01035419.1 between positions 17597 and 19143 in the reverse strand. The gene contains 2 exons but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish TR1 protein found in UniProt, because we could not find any information about it in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.

Protein TR1~B is located in contig number BADN01055795.1 between positions 1230 and 4182 in the forward strand. The gene contains 4 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish TR1 protein found in UniProt, because we could not find any information about it in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
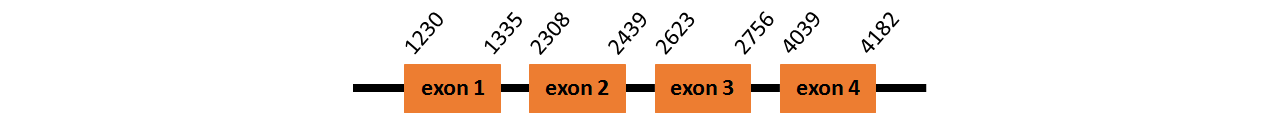
Protein TR1~C is located in contig number BADN01024054.1 between positions 3376 and 3588 in the forward strand. The gene contains 1 exon, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish TR1 protein found in UniProt, because we could not find any information about it in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found did not find any SECIS element in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.

Protein TR2 is located in contig number BADN01034111.1 between positions 1691 and 5342, in contig number BADN01034113.1 between positions 2458 and 3249 and in contig number BADN01034115.1 between positions 47 and 3467, all of them in the forward strand. The gene contains 20 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human TR2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found a SECIS element of grade A predicted in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
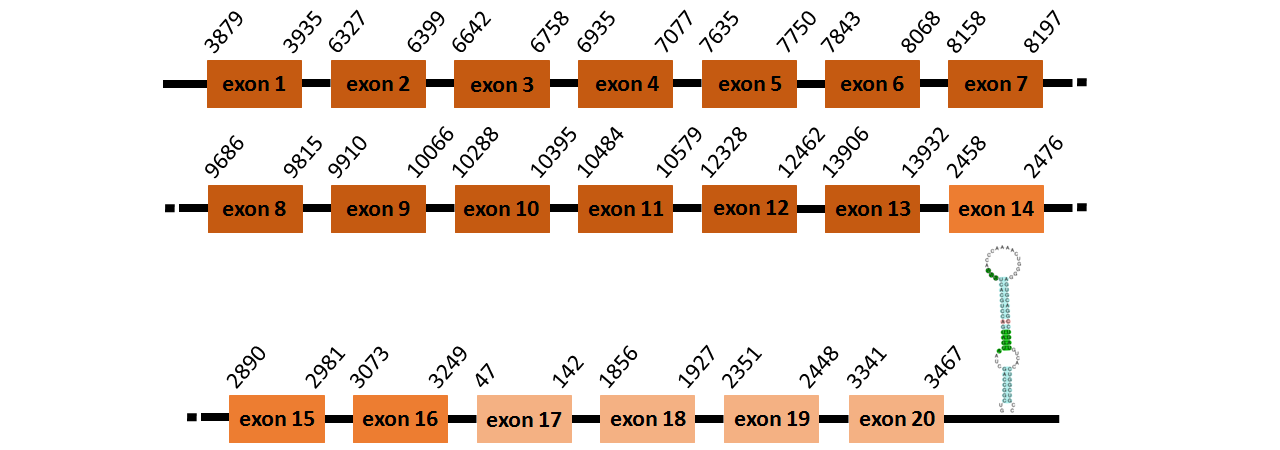
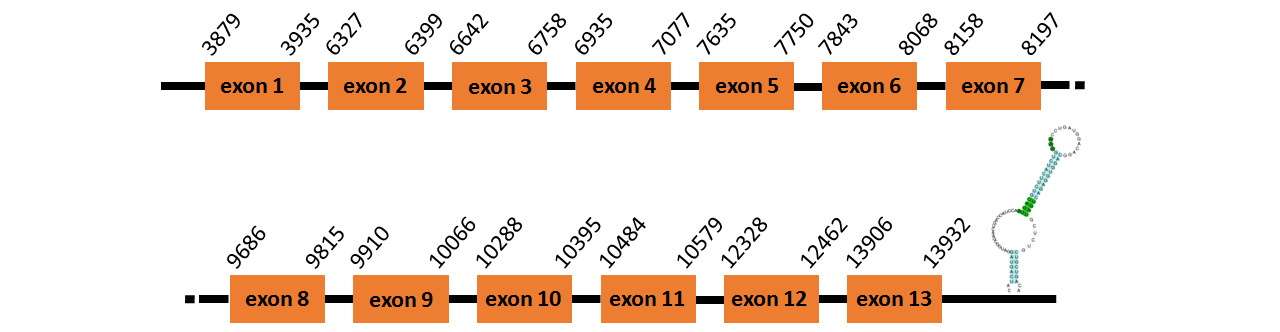
Protein TR3 is located in contig number BADN01057254.1 between positions 3879 and 13932 in the forward strand. The gene contains 13 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human TR3 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein eEFsec is located in contig number BADN01010202.1 between positions 9890 and 10168, in contig number BADN01010203.1 between positions 1203 and 2330 and in contig number BADN01010204.1 between positions 404 and 6182, all of them in the forward strand. The gene contains 7 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish eEFsec protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

Protein PSTK is located in contig number BADN01076497.1 between positions 1247 and 3572 in the forward strand. The gene contains 6 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the human PSTK protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

Protein SBP2 is located in contig number BADN01086679.1 between positions 1117 and 3097 in the forward strand. The gene contains 8 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the human SBP protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.

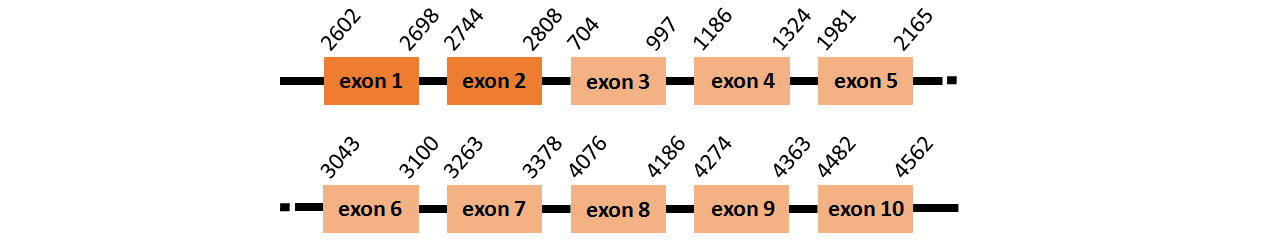
Protein SBP2L is located in contig number BADN01061252.1 between positions 2602 and 2808, and BADN01061253.1 between positions 704 and 4562, both of them in the forward strand. The gene contains 10 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SBP protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
Protein trnau1ap is located in contig number BADN01022579.1 between positions 10466 and 14347 in the forward strand. The gene contains 9 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SECp protein found in UniProt, because SelenoDB did not have this protein properly annotated. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
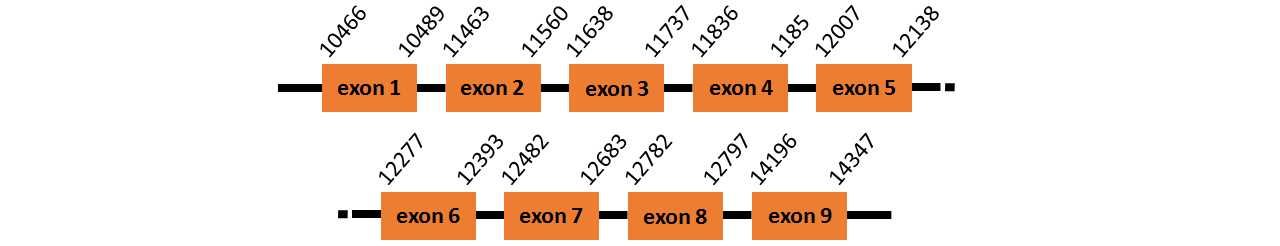
Protein trnau1ap like is located in contig number BADN01039777.1 between positions 13929 and 20064, and contig BADN01039778.1 between positions 475 and 609 in the forward strand. The gene contains 11 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SECp43 protein found in SelenoDB. To find the gene structure we analyzed the exonerate.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
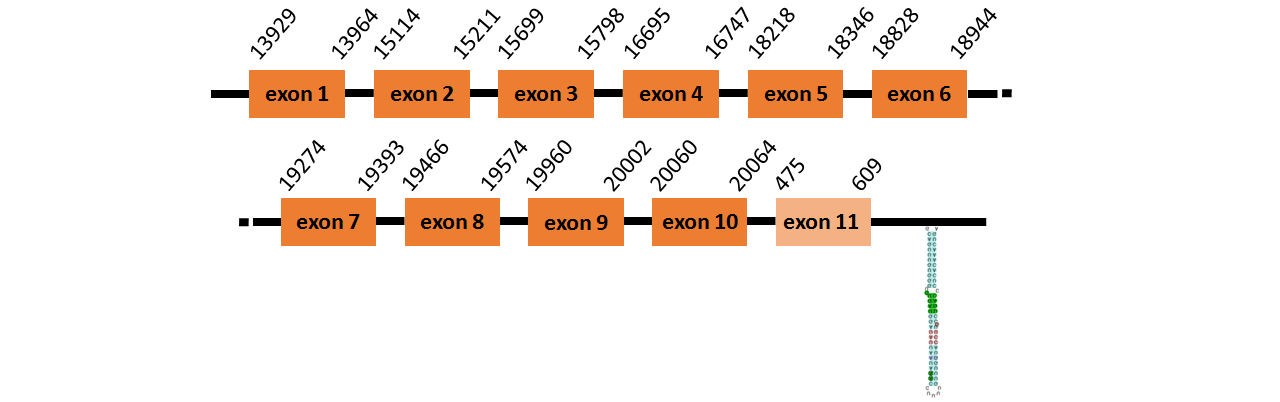
Protein trnau1ap like ~ is located in contig number BADN01065248.1 between positions 1759 and 1956 in the forward strand. The gene contains 1 exons, but it does not include any Selenocystein.
This protein did not have its correct name in SelenoDB, so we followed the steps defined in the Methodology in order to find its proper name. This protein was predicted blasting Thunnus orientalis genome against the zebrafish SECp43 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.

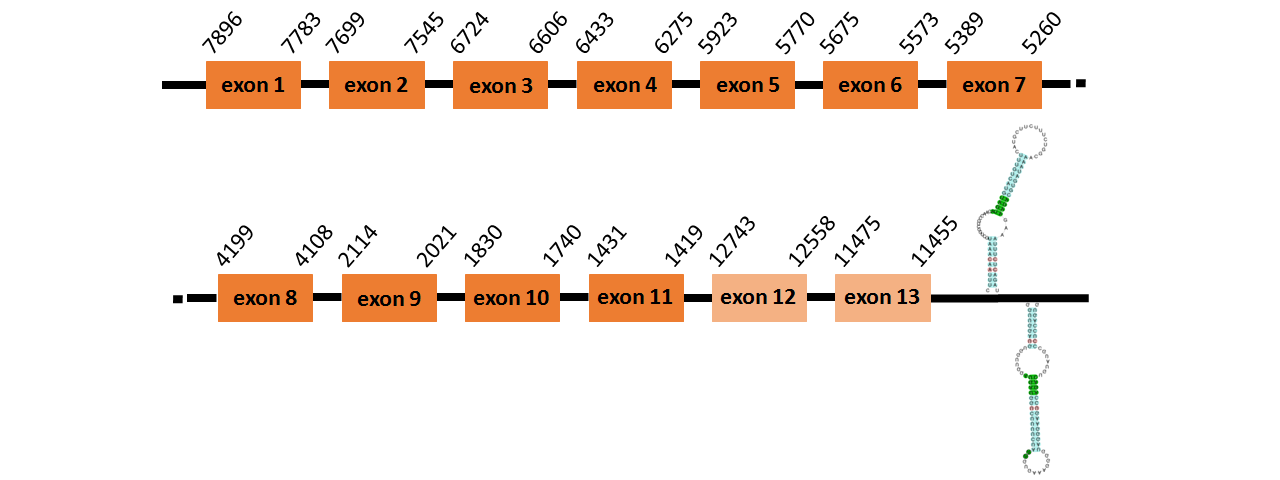
Protein SecS is located in contig number BADN01005541.1 between positions 11455 and 12743 and in contig number BADN01005542.1 between positions 1419 and 7896, both of them in the reverse strand. The gene contains 13 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SPS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We found two SECIS elements, both of them of grade B, in the 3’UTR region.
We obtained an output from Selenoprofiles that matches with our results about this protein.
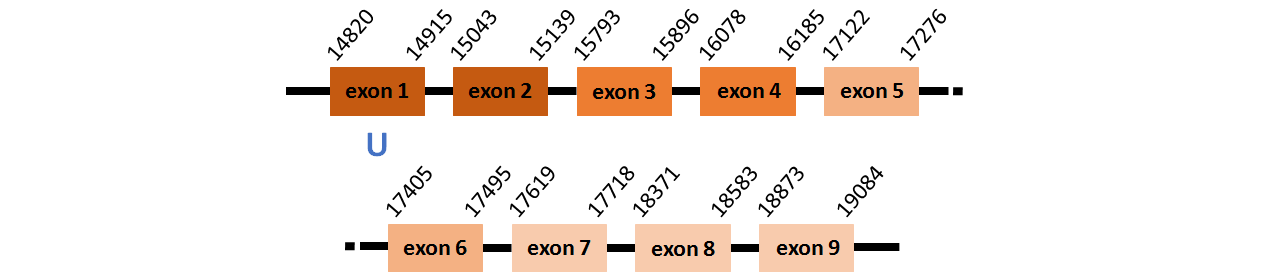
Protein SPS1 is located in contig number BADN01009667.1 between positions 14820 and 19084 in the forward strand. The gene contains 3 exons, but it does not include any Selenocystein.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SPS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We obtained an output from Selenoprofiles that matches with our results about this protein.
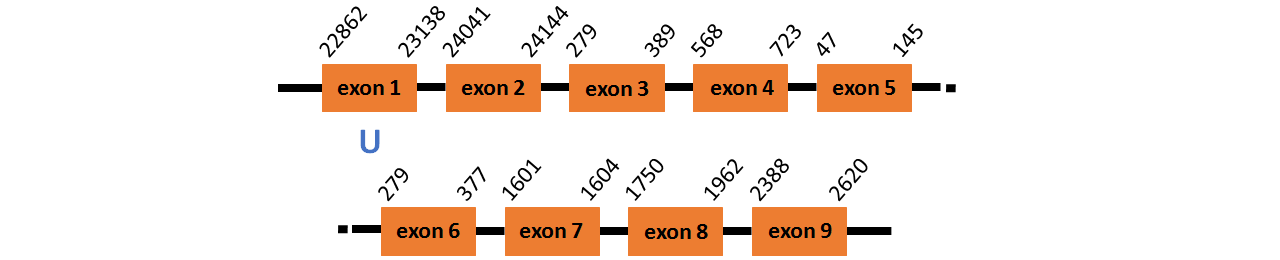
Protein SPS is located in contig number BADN01057937.1 between positions 22862 and 24144, contig BADN01057938.1 between positions 279 and 723, contig BADN01057939.1 and 47 and 37, and contig BADN01057941.1 between positions 1601 and 2620, all of them in the forward strand. The gene contains 9 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SPS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We did not find any SECIS elements in this protein.
We did not obtained any output from Selenoprofiles that matches with our results about this protein.
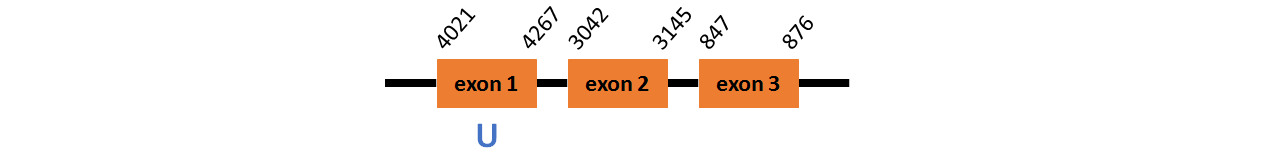
Protein SPS2~ is located in contig number BADN01117614.1 between positions 847 and 4267 in the forward strand. The gene contains 3 exons, with a Selenocystein in exon 1.
This protein was predicted blasting Thunnus orientalis genome against the zebrafish SPS2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate.
We did not find any SECIS elements in this protein.
We did not obtain any output from Selenoprofiles that matches with our results about this protein.
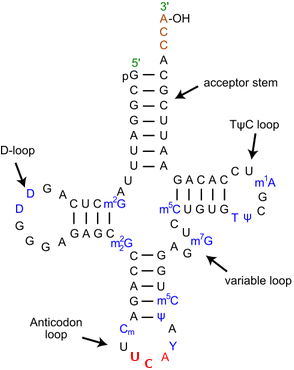
Result analysis of tRNA-Sec
Selenoproteins contain the amino acid Sec. During transcription Sec needs the presence of SECI elements and proteins that form part of the machinery, as well as the specific tRNA for this amino acid. To localize the tRNA-Sec in our genome we have used the programs known as Aragorn and tRNAscan-SE. After using Aragorn, we have found the localization of the following contigs:
BADN01026095.1
BADN01040258.1
BADN01048366.1
BADN01071448.1
BADN01087975.1
BADN01113814.1
BADN01132654.1
In comparison, while using the program tRNA-SE we have been able to find the following contigs:
BADN01040258.1
BADN01113814.1
BADN01132654.1
As the results show, we can assure that the contigs that contain the tRNA-Sec, those ones named above, are well predicted because they also appear as a result provided by Aragorn. The rest of the contigs that are stated in the output file when using Aragorn we are reticent to convincingly affirm if they are tRNA-Sec, but we could assume it is a positive result.