In order to identify all the selenoproteins and its machinery genes from Laticauda laticaudata we have studied the homology between the selenoproteins of Homo sapiens and the proteins encoded by the genes of L. laticaudata . Our findings are shown below.
Glutathione peroxidase is the general name for the largest family in vertebrates of multiple isozymes that catalyze the reduction of H2O2 or organic hydroperoxides to water or corresponding alcohols using reduced glutathione (GSH) as an electron donor (Margis R et al., 2008). Mammals have eight GPx homologs: GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7 and GPx8. GPx1-6 contain a Sec residue in their active site. In the other GPx homologs (GPx7 and GPx8), the active-site Sec is replaced by a Cys (Toppo S et al., 2008)(Brigelius-Flohé R et al., 2013). Among the GPx family, GPx1 is considered as the major enzyme responsible for removing H2O2 so it is a major antioxidant enzyme that protects cells against lethal oxidative stress (Avery JC et al., 2018).
Here we provide the phylogenetic tree of the human and L. laticaudata GPx family:
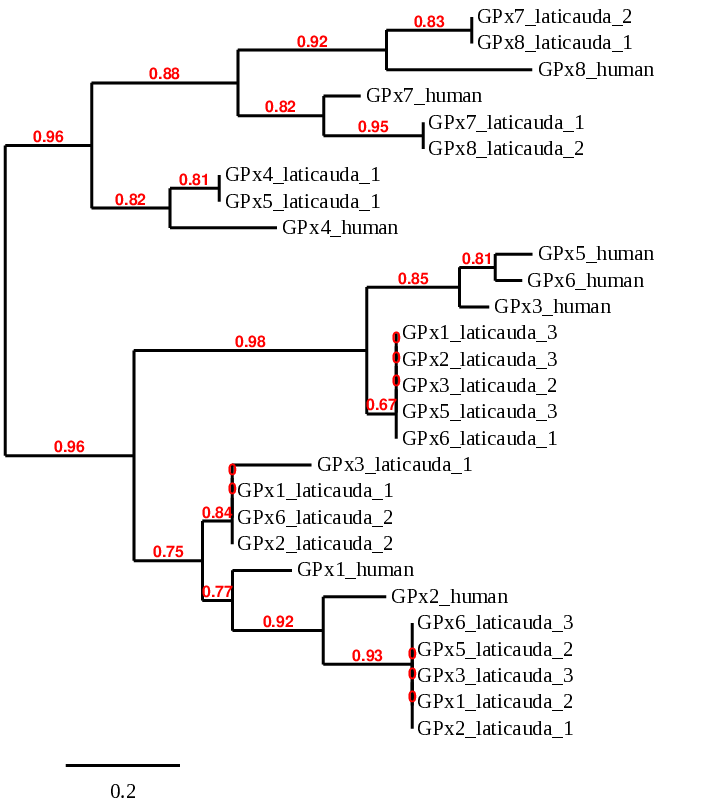
When aligning with the GPx1 protein from human, 3 scaffolds of the L. laticaudata GPx1 show significant hits based on our criteria: BHFT01025296.1 (GPx1_laticauda_1), BHFT01049178.1 (GPx1_laticauda_2) and BHFT01000450.1 (GPx1_laticauda_3). However, we can discard the second and the third prediction because they correspond to the human GPx2 and GPx3, respectively, due to their similarity. In the phylogenetic tree we can also see that in consequence of the similarity of the GPx proteins, other predictions are aligned with GPx1. So, we only have one good prediction in BHFT01025296.1 scaffold that has low E-value and high coverage.
We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 25466 between the positions 8179 and 10734 in the reverse strand (-). Two exons are predicted:
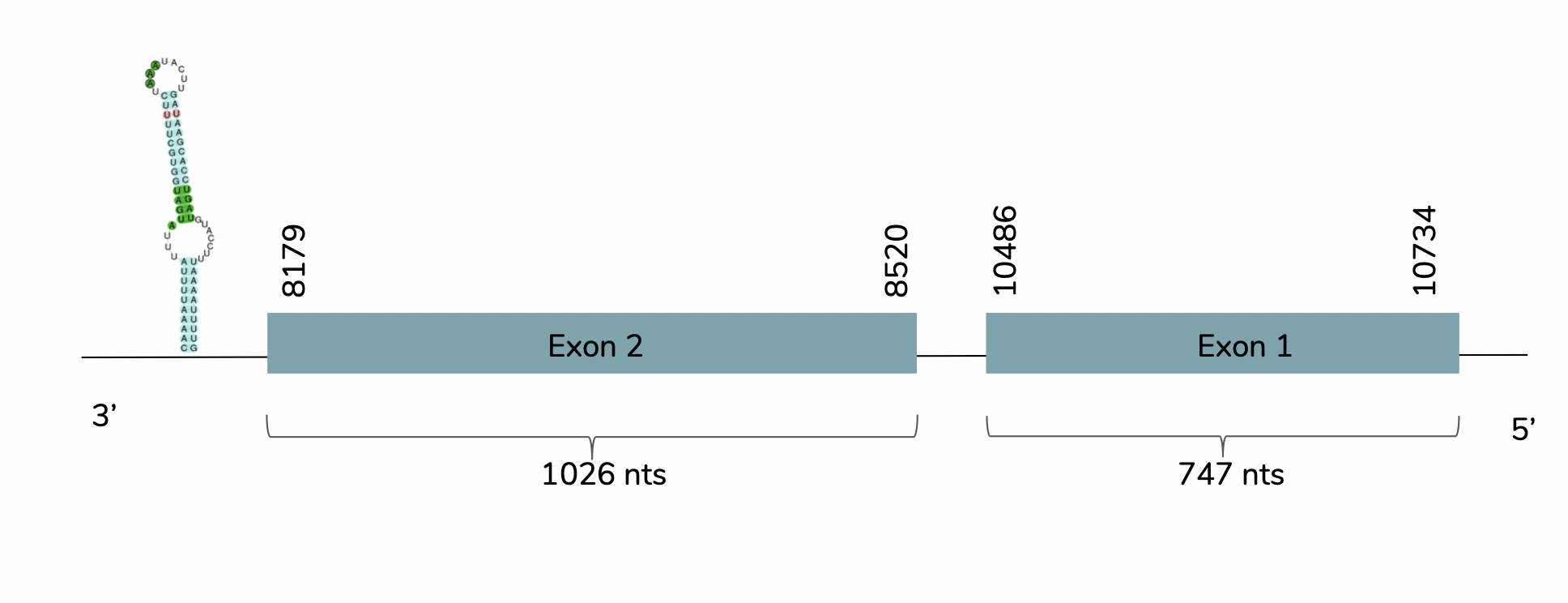
The T-coffee showed a very good alignment (score=992) between the predicted GPx1 and the human GPx1. The predicted protein has a Sec residue aligned with a Sec of the human GPx1 but does not include a Methionine (Met) at the beginning of the protein. However, when using Seblastian, a better prediction is obtained due to the fact that the Met is included. As it is showed above, one SECIS candidate is predicted at the 3'-UTR end between positions 7677 and 7610 in the reverse strand.
Taken all together, we can conclude that the GPx1 gene is found in the Laticauda laticaudata genome and corresponds to the human GPx1. We can also say that the predicted protein is a selenoprotein and an orthologous of the human GPx1.
In the case of GPx2, 3 scaffolds show significant hits when aligning with the GPx2 protein from human: BHFT01049178.1 (GPx2_laticauda_1), BHFT01025296.1 (GPx2_laticauda_2) and BHFT01000450.1 (GPx2_laticauda_3). However, the one that shows a lower E-value and a higher coverage is the one selected, BHFT01049178.1. In order to check that this is the most accurate prediction we performed a phylogenetic tree. So, we can confirm that the prediction selected is the one that shows the most similarity. As the GPx family shows high similarity, we find 4 other predictions that would correspond to the GPx2 gene.
As we can observe with Exonerate the gene is found in the Laticauda laticaudata genome in contig 497 and 2 exons are predicted between the positions 17974 and 21709 in the forward strand (+):
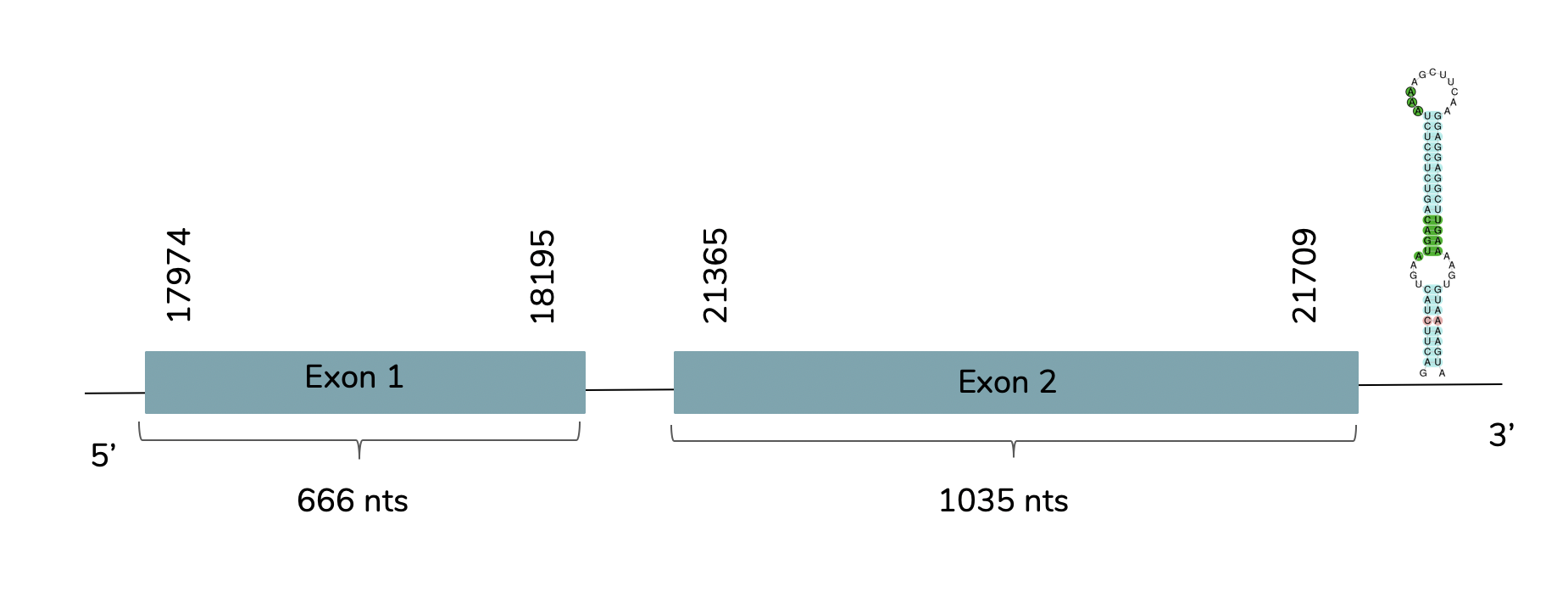
T-coffee supports the hypothesis that this prediction corresponds to GPx2. Also we can observe that the predicted protein has a Sec residue. Using Seblastian one grade A SECIS candidate is predicted at the 3'-UTR end between positions 21912-21976 in the forward strand (+).
We conclude that the gene corresponds to GPx2, that it is found in the Laticauda laticaudata genome and that it is a selenoprotein. Finally, we can say that human GPx2 and our GPx2 predicted are orthologous.
In this case, we obtain 3 scaffolds with significant hits when aligning with the GPx3 protein from human: BHFT01025296.1 (GPx3_laticauda_1), BHFT01000450.1 (GPx3_laticauda_2) and BHFT01049178.1 (GPx3_laticauda_3). The BHFT01000450.1 scaffold is the one studied because it shows a lower E-value and a higher coverage than the others. We performed a phylogenetic tree in order to check that this is the most accurate and similar to the human GPx3. We can see that the prediction selected (GPx3_laticauda_2), that is located in the scaffold BHFT01000450.1 is the most accurate. As the GPx family shows high similarity, we find 4 other predictions that would correspond to the GPx3 protein, specifically between the positions 23000 and 27000.
We conclude with Exonerate that the gene is found in the L. laticaudata genome in contig 449 and 5 exons are predicted between the positions 18185 and 27852 in the forward strand (+):
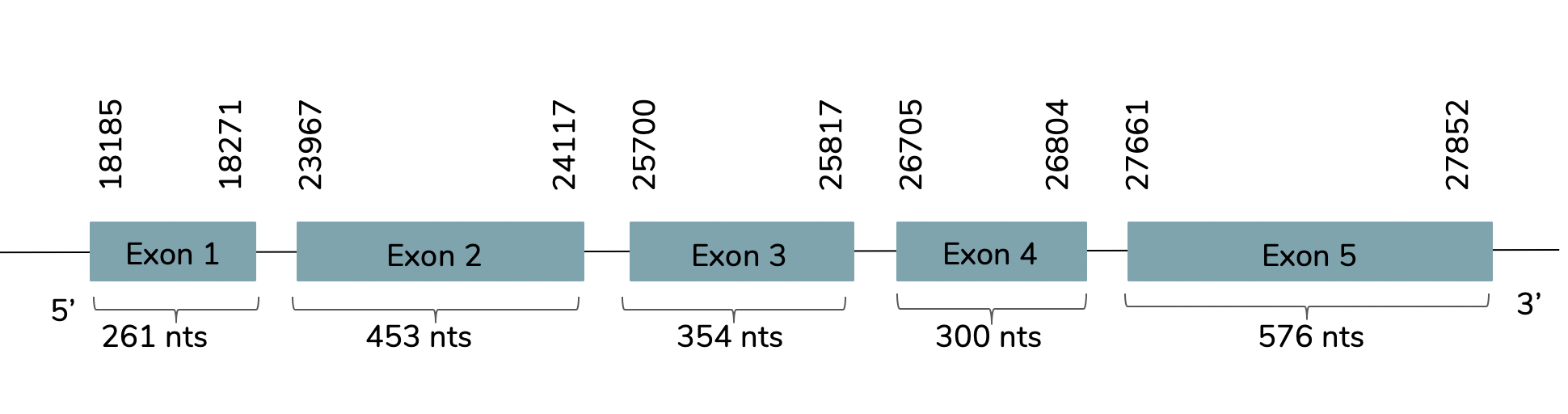
The T-coffee showed a value around 1000 (score=977) and it also showed that it is a good prediction. Moreover, in the predicted protein we can find a selenocysteine aligned with the human GPx3 and it includes a Met at its beginning. However, Seblastian does not predict any selenoprotein inside the L. laticaudata sequence studied. Although two SECIS are predicted, the two of them are rejected. One of them is rejected because it is not in the same strand of the predicted gene and the other one is discarded because it is predicted before the end of our gene (23661-23736).
So, we can conclude that the gene corresponds to GPx3 and that it is found in the L. laticaudata genome, but we can not conclude that the GPx3 predicted is a selenoprotein because it does not have a SECIS element.
For GPx4, we find two possible hits in BHFT01021643.1 and BHFT01025296.1 scaffolds with the tBLASTn. However, with Exonerate we can discard the hit found in BHFT01025296.1 scaffold because no prediction is made. So, we only have one good prediction in the scaffold BHFT01021643.1 (GPx4_laticauda_1) with high coverage percentage and a low e-value.
Exonerate shows that the gene is found in the Laticauda laticaudata genome in contig 21755 and that it has 3 exons between the positions 74379 and 76696 in the reverse strand (-):
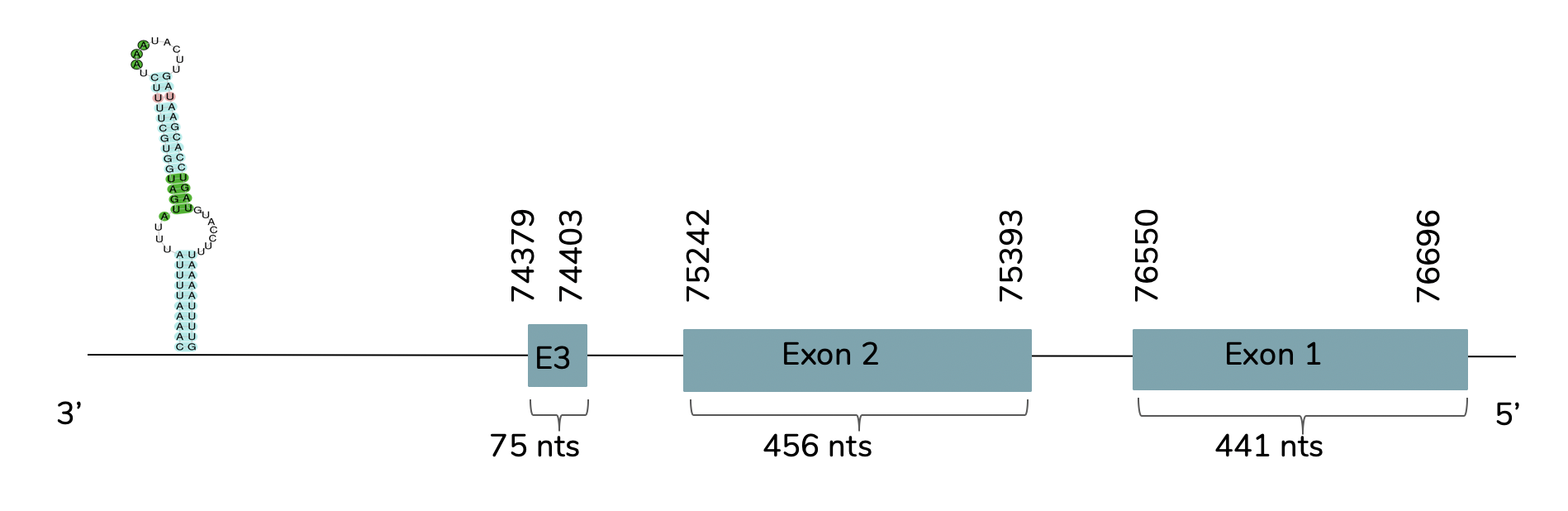
The T-coffee shows a good prediction and also a very good alignment between the human GPx4 and the predicted one. This prediction contains a Sec residue aligned with the human GPx4 but does not include a Met at the beginning of the protein. With Seblastian, we obtain a better prediction due to the fact that shows more aminoacids in the C-terminal and N-terminal. Also, one SECIS candidate is predicted at the 3'-UTR end between positions 70507 and 70432 in the reverse strand.
In conclusion, we can say that the gene GPx4 is a selenoprotein and that it is found in the L. laticaudata genome. As it is showed in the phylogenetic tree, we can also say that the human GPx4 and the predicted GPx4 are orthologous.
We find 3 scaffolds of GPx5 protein that showed significant hits when aligning with the GPx5 protein from human: BHFT01025296.1 (GPx5_laticauda_1), BHFT01049178.1 (GPx5_laticauda_2) and BHFT01000450.1 (GPx5_laticauda_3). However, we can say that this protein is lost in the Laticauda laticaudata genome because all the predictions correspond to others GPx proteins due to their similarity (see the phylogenetic tree ).
In the case of GPx6, 3 scaffolds showed significant hits when aligning with the human GPx6 protein: BHFT01000450.1 (GPx6_laticauda_1), BHFT01025296.1 (GPx6_laticauda_2) and BHFT01049178.1 (GPx6_laticauda_3). However, as well as for GPx5, we can say that the Laticauda laticaudata genome does not contain this protein because as we can see in the phylogenetic tree all the predictions correspond to others GPx proteins due to their similarity.
When aligning with the GPx7 protein from human, we find 2 significant hits in the BHFT01060761.1 (GPx7_laticauda_1) and BHFT01032132.1 (GPx7_laticauda_2) scaffolds. However, the one that showed a lower E-value and a higher coverage was the one selected, BHFT01060761.1. In order to check which of the predictions was the most accurate and showed more similarity with the human GPx7, we performed a phylogenetic tree. In the tree we can observe that due to the similarity between GPx7 and GPx8, a prediction of GPx8 corresponds to human GPx7.
We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 61691 and 3 exons are predicted. The gene predicted is located between the positions 28537 and 34666 in the reverse strand (-):
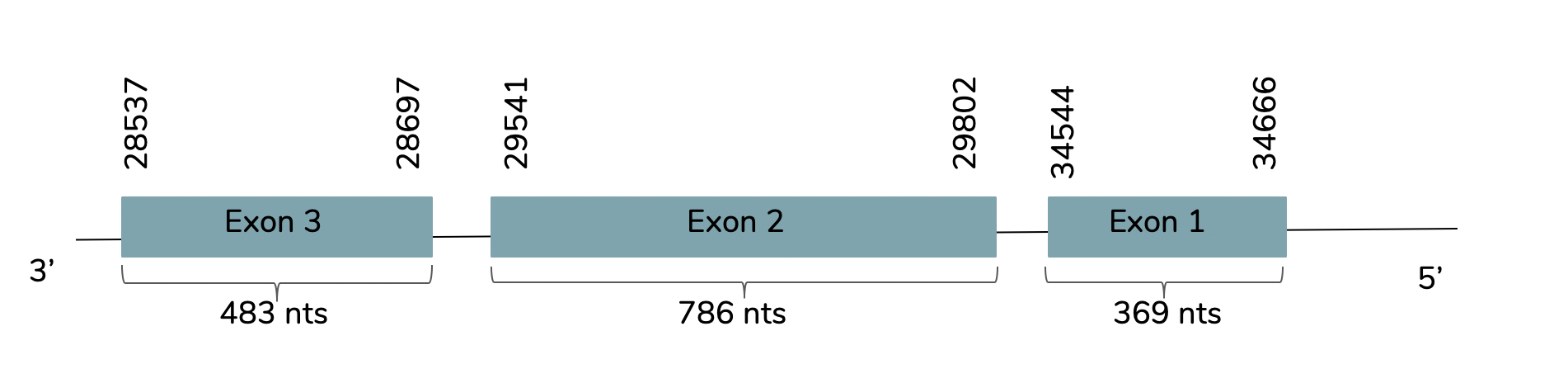
The T-coffee showed a value around 1000 and it also showed that it is a good prediction. However, the predicted protein does not include a Met at the beginning of the protein and as human GPx7 the Sec residue is replaced by Cys. Moreover, Seblastian do not predict any protein or SECIS elements.
So, we can say that this protein is a cysteine-containing homolog, same as the human one. Also, we can say that the predicted GPx7 protein is found in the Laticauda laticaudata genome corresponding with the human GPx7.
In the case of GPx8, 2 scaffolds showed significant hits when aligning with the GPx8 protein from human: BHFT01032132.1 (GPx8_laticauda_1) and BHFT01060761.1 (GPx8_laticauda_2). However, the one that showed a lower E-value and a higher coverage was the one selected, BHFT01032132.1. We performed a phylogenetic tree in order to ensure that this is the most accurate prediction and the one that showed more similarity with the human GPx8. In the tree we can also observe that due to the similarity between GPx7 and GPx8, a prediction of GPx8 corresponds to human GPx7.
With Exonerate we conclude that the gene studied is found in the Laticauda laticaudata genome in contig 32395 and located between the positions 19841 and 21106 in the forward strand (+). In this gene two exons are predicted:
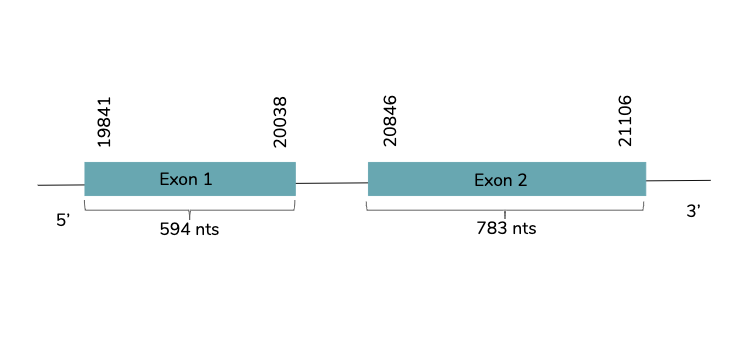
T-coffee showed a very good alignment with a score of 1000. It also showed that it is a good prediction but the predicted protein does not include a Met at the beginning of the protein nor a selenocysteine residue. As human GPx8 the Sec residue is replaced by Cys. In the sequence studied, no SECIS are found. Also, Seblastian do not find any protein in the analysed sequence.
In conclusion, this protein is a cysteine-containing homolog and we can say that the predicted GPx8 is found in the Laticauda laticaudata genome corresponding with the human GPx8.
Iodothyronine deiodinases (DI) are important mediators of thyroid hormone (TH) action. Three different types of iodothyronine deiodinases (D1, D2 and D3) have been identified in vertebrates from fish to mammals. They share several common characteristics, including a selenocysteine residue in their catalytic centre. The three types of DI belong to the thioredoxin fold superfamily and share a similar structural organisation, but show also some type-specific difference. These specific characteristics seem very well conserved for D2 and D3, while D1 shows more evolutionary diversity (Daras VM et al., 2012). The active-site Sec residue is located in the N-terminal part of the protein. However, in DI2, an additional Sec, whose function is unknown, is present in the C-terminal region (Mariotti M et al., 2012).
Here we provide the phylogenetic tree of the human and L. laticaudata DI family:
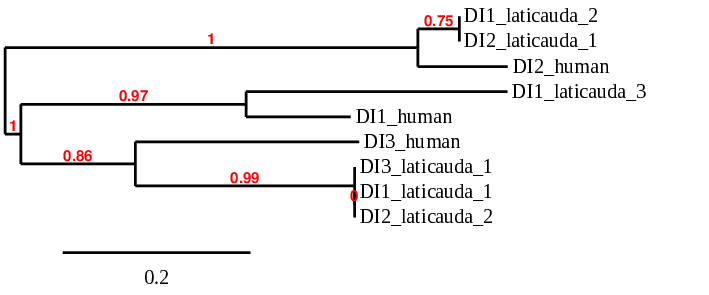
When aligning with the DI1 protein from human, 3 scaffolds showed significant hits: BHFT01042446.1 (DI1_laticauda_1), BHFT01040576.1 (DI1_laticauda_2) and BHFT01004800.1 (DI1_laticauda_3). However, as it is shown in the phylogenetic tree, the one that showed more similarity with the DI1 protein from human (DI1_human) was the one selected: BHFT01004800.1 (DI1_laticauda_3). We conclude with Exonerate that the gene is found in Laticauda laticaudata genome in contig 4799 between the positions 77633 and 80803 in the forward strand (+) and 3 exons are predicted:
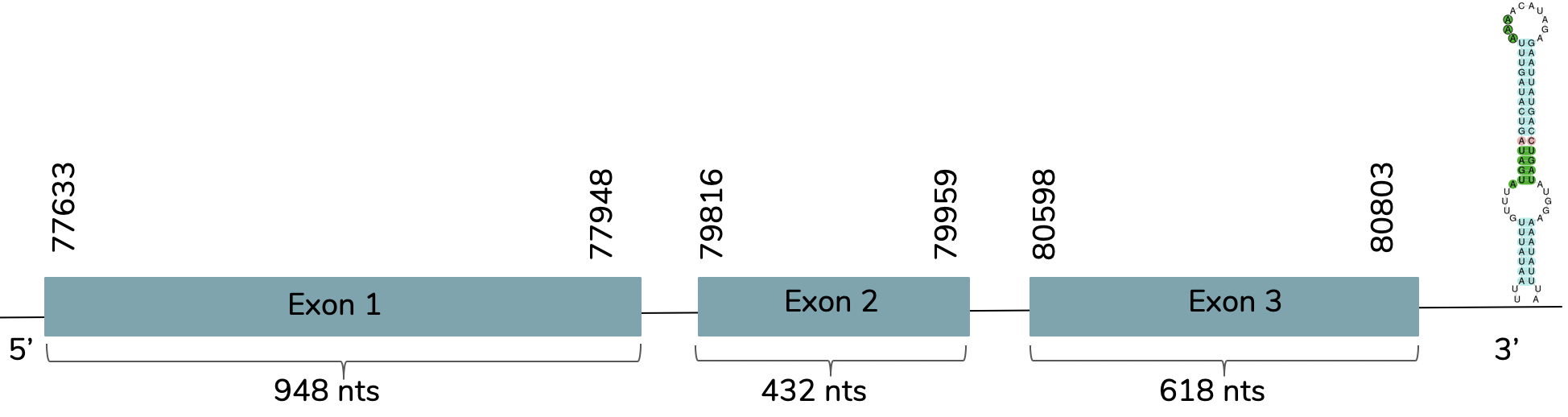
We obtained a very good alignment (score of 996) between the predicted DI1 and the human query in T-coffee and the predicted protein in L.laticaudata has a Sec residue. We also observed that the predicted DI1 does not include a Met residue at the beginning of the protein. This can be because exonerate has not correctly predicted the structure of the gene. With Seblastian we obtained a better prediction due to the fact that shows more aminoacids in the C-terminal and the N-terminal. Also, one SECIS candidate is predicted at the 3'-UTR end between positions 82160 and 82227 in the forward strand (+). We must say that the fasta subseq of the protein was too large, so we did the fasta subseq command again in order to make it shorter.
Taken all together, we conclude that this protein is a selenoprotein and we can say that the predicted DI1 is found in the Laticauda laticaudata genome corresponding with the human DI1. Moreover, the human and the predicted DI1 are orthologous.
For DI2, 2 scaffolds showed significant hits when aligning with the DI2 protein from human: BHFT01040576.1 (DI2_laticauda_1) and BHFT01042446.1 (DI2_laticauda_2). The scaffold that showed more similarity with the DI2 protein from human (DI2_human) was the one selected: BHFT01040576.1 (DI2_laticauda_1). In order to check that this is the most accurate prediction and the one that shows more similarity with the human DI2, we performed a phylogenetic tree. In the tree we can also observe that due to the family similarity, we can find predictions of the other DI proteins corresponding to human DI2.
Exonerate concludes the presence of the gene of DI2 in the genome of Laticauda laticaudata in contig 40972 in the reverse strand (-) between the positions 8942 and 24344. Two exons are predicted:
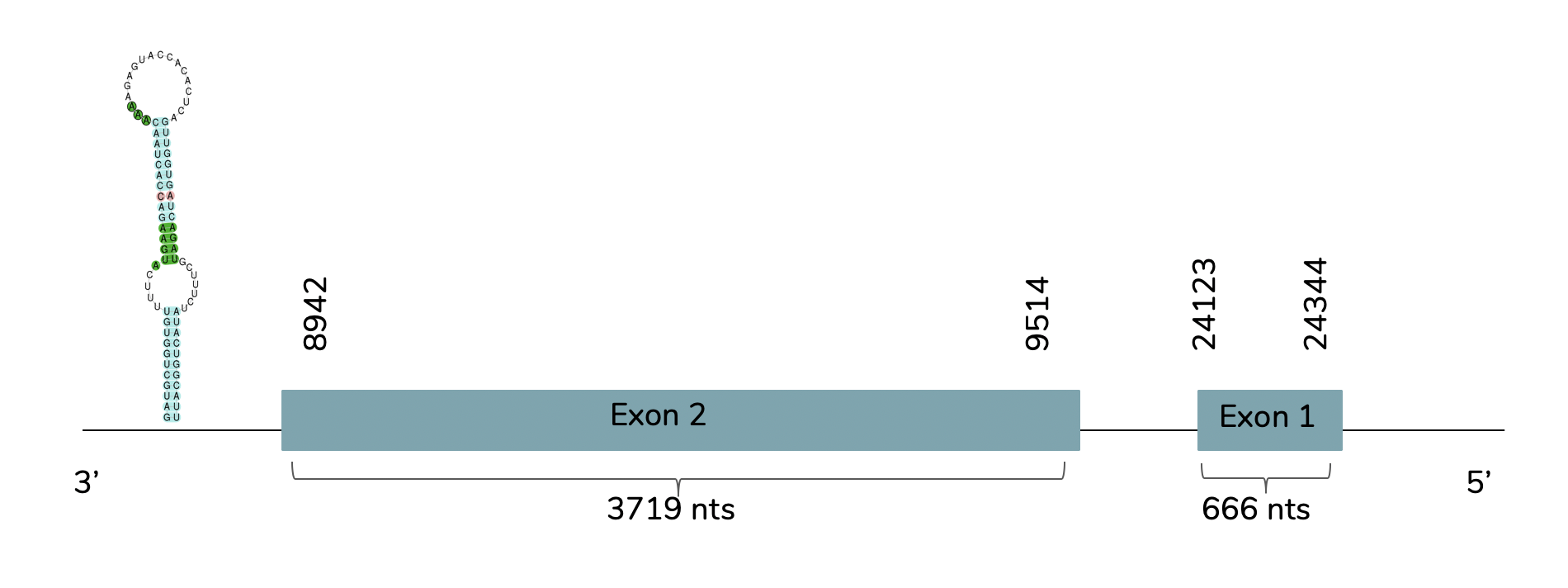
In this case, the predicted DI2 also show a very good alignment with the human query in T-coffee (score of 979) and it has a Sec residue aligned with the human DI2. Also, the predicted protein includes a Met at the beginning of the protein and Seblastian predicted DI2 inside the studied L. laticaudata sequence. With Seblastian we obtained a better prediction due to the fact that shows more aminoacids in the N-terminal. One SECIS candidate with grade A is predicted at the 3'-UTR end between positions 7547 and 7470 in the reverse strand (-).
We can conclude that this protein is a selenoprotein and that the predicted DI2 is found in the Laticauda laticaudata genome corresponding with the human DI2. Moreover, the human and the predicted DI2 are orthologous.
We got only one good prediction in the scaffold BHFT01042446.1 (DI3_laticauda_1) with a high identity percentage when aligning with the DI3 protein from human. We performed a phylogeny tree in order to check that this prediction is the one that shows more similarity with the human DI3. We can also see predictions of the other DI proteins corresponding to human DI3, due to the family similarity.
With Exonerate we observe that this prediction has only one exon that goes from the position 30323 to the 30958 in contig 42883 in the forward strand (+):
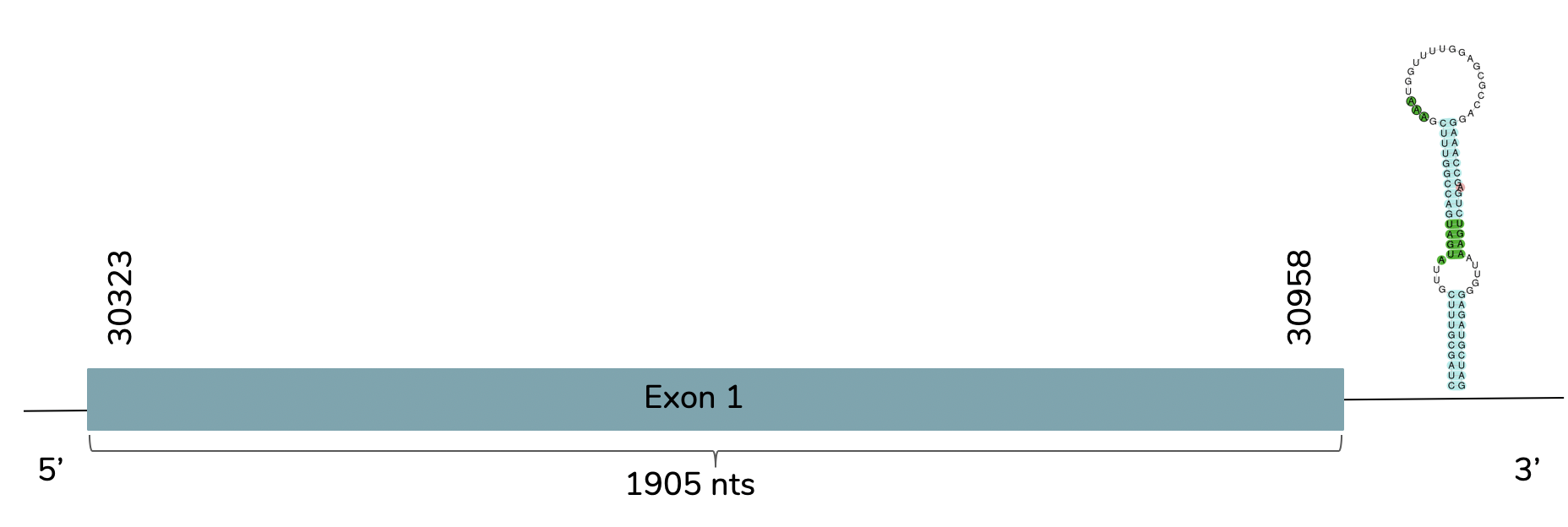
The T-coffee shows a score of 994 and that this is a very good alignment. A Sec residue is found in the predicted protein of L. laticaudata and it does not include a Met at the beginning. Using Seblastian a better prediction is showed due to the fact that shows more aminoacids in the C-terminal. One SECIS candidate with grade A is predicted at the 3'-UTR end between positions 31464 and 31542 in the forward strand (+).
To conclude, this protein is a selenoprotein found in the L. laticaudata genome and presents orthology with human DI3.
Selenoprotein M (SelM) and Selenoprotein 15 (Sel15) are 15 kDa proteins that share 31% sequence identity and localize to the ER. SelM and Sel15 have Cys-X-X-Sec and Cys-X-Sec motifs, respectively. UGTR is involved in the quality control of protein folding, and both proteins have been suggested to play a role in protein-folding in the ER (Revees MA et al., 2009).
In this case, we got only one prediction in the scaffold BHFT01039670.1 and using tBLASTn we find that it has low E-value but also low query coverage percentage. Exonerate shows a better coverage and looking at the prediction we find that the gene is found in contig 40047 and it has 2 exons between the positions 34393 and 35460 in the reverse strand (-):

The T-coffee showed that the alignment is mostly good. This prediction does not contains a Sec residue nor includes a Met at the beginning of the protein. Seblastian find a better protein prediction because it contains more aminoacids in the C-terminal. A SECIS element is found in the 3’UTR between the positions 34009 and 33942 in the reverse strand.
So, we can conclude that the gene corresponds to SelM and that it is found in the Laticauda laticaudata genome. However, we can not ensure that SelM is a selenoprotein because it does not have a selenocysteine residue, because the genome of L. laticaudata is fragmented or because the human and the L. laticaudata proteins have diverged too much in order to predict the residue.
In this case, we found two relevant hits in BHFT01037660.1 (Sel15_laticauda_1) and BHFT01072119.1 (Sel15_laticauda_2) scaffolds. Looking at the start and end percentages of query (23% to 68% and 67% to 84%), we hypothesized that it is a truncated protein in different scaffolds. We intend to join the gff documents of the two proteins and then, make a fastafetch but, looking at the percentages, they overlap, so a frameshift would be created. Taking this into account, we have checked the results of the two scaffolds in order to see if the protein is indeed truncated.
Using Exonerate, we find that the first part of the protein (Sel15_laticauda_1) has one gene in contig 37995 and that it has 1 exon between the positions 27256 and 27426 in the forward strand (+). We also performerd Exonerate in the second part of the protein (Sel15_laticauda_2) and it finds the gene in contig 73434 with 2 exons between the positions 3472 and 10778 in the forward strand (+). This prediction contains a Sec residue in the end part of the protein (Sel15_laticauda_2) but does not include a Met at the beggining of the protein:

A better prediction of Sel15_laticauda_2 is made using Seblastian because it contains more aminoacid in the C-terminal. We also find a SECIS element in the 3'UTR end between positions 11469 and 11542 in the forward strand (+). So, we can conclude that the protein is truncated in two different scaffold and that it is a selenoprotein. Thus, the predicted Sel15 is found in the Laticauda laticaudata genome.
The Selenoprotein I (SelI) is a membrane protein that possesses a CDP-alcohol phosphatidyltransferase motif, a common motif conserved in phospholipid synthases. It plays a central role in maintenance of vesicular membranes (Horibata Y et al., 2007).
In this case, we got only one good prediction in the scaffold BHFT01018958.1 with low e-value and high query coverage percentage. Exonerate shows us that the gene is found in contig 19028 and 9 exons are predicted between the positions 1698 and 30398 in the forward strand (+):

The T-coffee showed a good alignment and a value around 1000. This prediction contains a Sec residue but does not include a Met at the beginning of the protein. Seblastian found a similar prediction and a SECIS element in the 3'UTR end between positions 31372 and 31451 in the forward strand (+).
We can, therefore, conclude that SelI is a selenoprotein found in L. laticaudata genome and that it is orthologous to the human SelI.
Selenoprotein K (SelK) is a transmembrane protein localized to the endoplasmic reticulum and involved in calcium flux in immune cells and ER associated degradation in cell lines (Verma S et al., 2011).
In this case, we got only one good prediction in the scaffold BHFT01021730.1 with low e-value but no high query coverage looking in the tBLASTn results. However, Exonerate shows high query coverage and founds a gene in contig 21844 with 4 exons predicted between the positions 62746-65818 in the reverse strand (-):

The T-coffee shows a very good alignment and a score around 1000. This prediction does not contains a Sec residue but includes a Met at the beginning of the protein. A very similar prediction is found using Seblastian and one SECIS element is predicted in the 3'UTR end between positions 61556 and 61633 in the reverse strand (-).
In conclusion, the SelK predicted is found in the L. laticaudata genome and it is orthologous to the human protein. However, we can not say that it is a selenoprotein because we can not find a Sec residue. This could be because of the fragmentation of the L. laticaudata genome or because the human region containing the residue is lost in our species protein.
Selenoprotein N (SelN) is involved in muscle development and maintenance. It also contributes in the regulation of oxidative stress and calcium homeostasis. It is located in the membrane of endoplasmic reticulum (Lescure A et al., 2009). Its deficiency causes several inherited neuromuscular disorders (SEPN1-related myopathies) (Castets P et al., 2012).
In this case, we got only one prediction in the scaffold BHFT01034693.1 with very high query coverage percentage and low e-value. We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 34988 and it has 11 exons and 10 introns between the positions 86313 and 102240 in the forward strand (+):
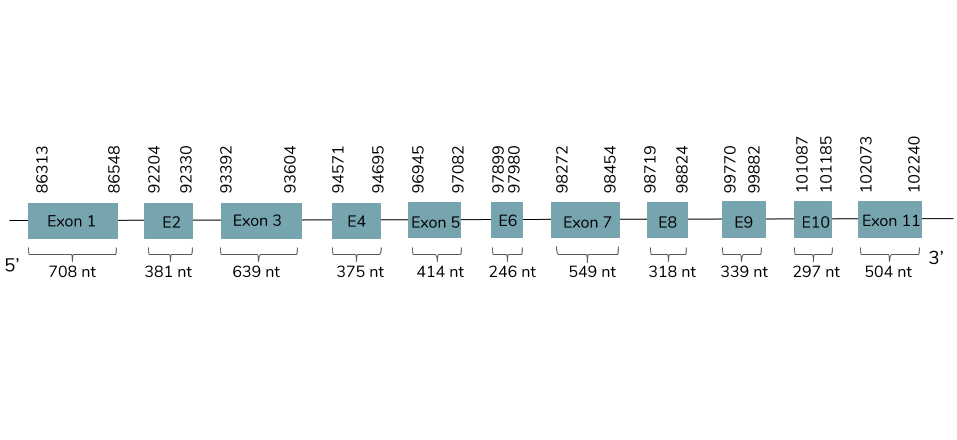
The T-coffee shows a very good alignment and a score around 1000. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Using Seblastian a very similar prediction is found and a SECIS element is predicted in the 3’UTR end between the positions 107027-107099 in the forward strand.
Taking this into account, we can conclude that SelN is found in the Laticauda laticaudata genome and that it is orthologous to the human SelN. We can also say that the predicted protein is a selenoprotein because of the presence of a Sec residue and a SECIS element. Comparing it with the human SelN, there has also occurred a change of a selenocysteine for a cysteine.
Selenoprotein O (SelO) is the largest mammalian selenoprotein with orthologs found in a wide range of organisms, including bacteria and yeast. SelO transfers AMP from ATP to Ser, Thr, and Tyr residues on protein substrates (AMPylation), uncovering their previously unrecognized activity (Screelatha A et al., 2018). It is suggested that SelO may be a redox-active mitochondrial selenoprotein which interacts with a redox target protein (Han SJ et al., 2014).
In this case, we got only one good prediction in the scaffold BHFT01043593.1 with high coverage percentage and low e-value. We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 44050 and it has 10 exons between the positions 12948 and 22955 in the forward strand (+):
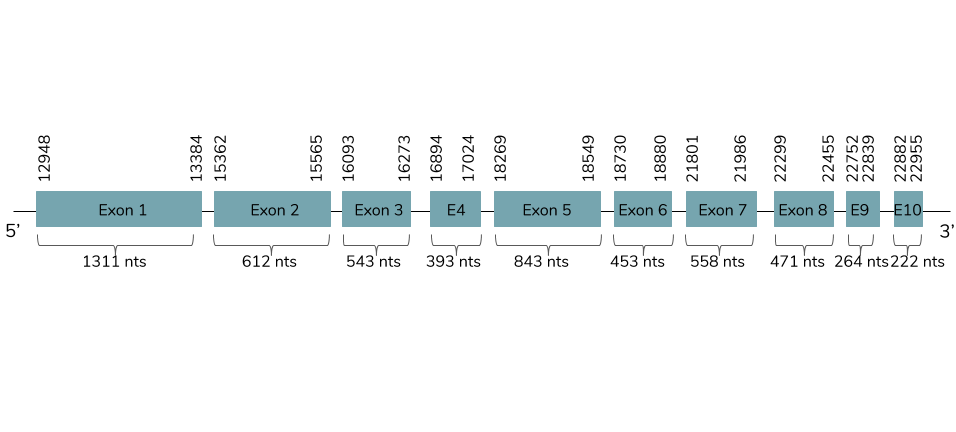
The T-coffee shows a value around 1000 and a good alignment. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Seblastian predicted a better alignment because it has more aminoacids in the N-terminal and it also has a Met in the beginning of the sequence. One SECIS element is predicted in the 3’UTR between the positions 23230 and 23308 in the forward strand.
So, we can conclude that the gene corresponds to SelO and that it is found in the Laticauda laticaudata genome. Moreover, the human SelO protein and the predicted one share an homologous relationship. We can also say that the predicted SelO is a selenoprotein, as we expected.
Selenoprotein P (SelP) is a secreted glycoprotein that contains most of the selenium in plasma (accounts for approximately 50% of the total Se of the plasma) (Burk RF et al., 2015). SelP homologs are found predominantly in vertebrates. The unique feature of SelP is the presence of multiple Sec residues. It is suggested that SelP plays an important role as an antioxidant and also in transporting Se to peripheral tissues (Labunskyy VM et al., 2014).
In the case of SelP, 2 scaffolds showed significant hits when aligning with the SelP protein from human: BHFT01045694.1 (SelP_laticauda_1) and BHFT01060482.1(SelP_laticauda_2). In order to check which of the predictions was the most accurate and showed more similarity with the human SelP, we performed the following phylogenetic tree:
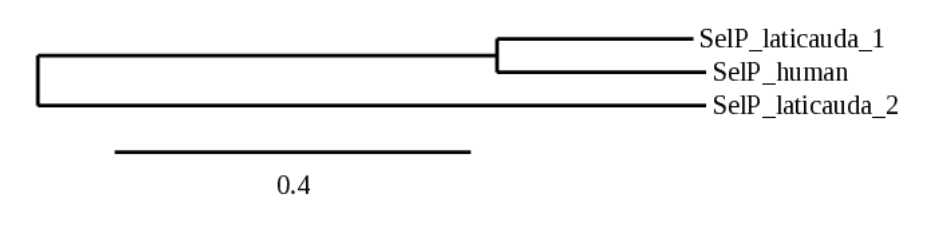
So, we select the scaffold BHFT01045694.1. However, the alignment of SelP_laticauda_1 only represents the first part of the protein. Maybe, the part of the end has lost in our specie. Using Exonerate, we find that the predicted alignment is in contig 46194 and contains 3 exons between the positions 18567 and 21028 in the forward strand (+):

The T-coffee shows a value around 1000 and a good alignment, although it corresponds only in the first part. This prediction contains a selenocysteine residue and includes a Met at the beginning of the protein. Seblastian predicted a better alignment because it has more aminoacids in the C-terminal. One SECIS element is predicted in the 3’UTR end between the positions 24194 and 24261 in the forward strand (+).
So, we can conclude that the gene corresponds to SelP and that it is found in the L. laticaudata genome, but the end part of it has lost. Moreover, the human SelP protein and the predicted one share an homologous relationship. We can also say that the predicted SelP is a selenoprotein, as we expected, but the human one contains 10 Sec residues that maybe has lost in our specie.
Methionine is a highly susceptible amino acid that can be oxidized to S and R diastereomeric forms of methionine sulfoxide by many of the reactive oxygen species generated in biological systems. Methionine sulfoxide reductases (Msrs) are thioredoxin-linked enzymes involved in the enzymatic conversion of methionine sulfoxide to methionine. Methionine Sulfoxide Reductase A (MsrA) is a sulfoxide reductase present in humans and some eukaryotic sepecies. It has been localized in different ocular regions and is abundantly expressed in the retina and in retinal pigment epithelial (RPE) cells. MsrA protects cells from oxidative stress (Sreekumar PG et al., 2011).
We decided to study only one prediction for MsrA gene. This prediction is located in the scaffold BHFT01047624.1 and has a high coverage percentage and low E-value. Exonerate shows that the gene is found in the L. laticaudata genome in contig 48178 and that it has two exons in the forward strand (+):

The T-coffee shows a value around 1000 and that it is a good prediction. The predicted protein contains a Sec residue and does not include a Met at the beginning of the protein. Using Seblastian, any SECIS elements are found nor a protein prediction. So, although it has a Sec residue we can not say that it is a selenoprotein. However, comparing it with the human MsrA, the predicted protein has gained a selenocysteine residue. Taking everything into account, the predicted MsrA is found in the L. laticaudata genome.
Methionine-R-Sulfoxide reductases are zinc-containing selenoproteins that were previously identified as Selenoproteins R (SelR) and Selenoprotein X (SelX). These proteins were found to function as a stereospecific methionine-R-sulfoxide reductases, which catalyzes repair of the R-enantiomer of oxidized methionine residues in proteins.
Here, we show the phylogenetic tree of SelR proteins from human and Laticauda laticaudata:
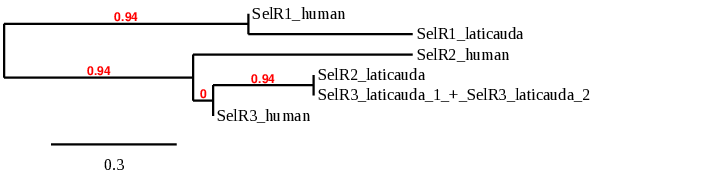
In this case, we got only one good prediction in the scaffold BHFT01066860.1 with high query coverage percentage and low e-value. We conclude with Exonerate that the gene is found in contig 67973 and it has 2 exons between the positions 5298 and 6451 in the reverse strand (-):

The T-coffee shows a very good alignment and a value around 1000. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Using Seblastian, a better prediction is made because the sequence starts with a Met and it has more aminoacids in the C-terminal. A SECIS element is predicted in the 3’UTR end between the positions 2636 and 2561 in the reverse strand (-).
In conclusion, the predicted SelR1 is found in the L. laticaudata genome and shares orthology with the human SelR1. Moreover, the L. laticaudata protein is a selenoprotein due to the presence of the Sec residue and a SECIS element.
In this case, we got 2 predictions in the scaffolds BHFT01052183.1 and BHFT01057691.1. However, the BHFT01057691.1 can be discarded due to the fact that Exonerate do not predict the protein in this scaffold. So, we only have one possible alignment. Exonerate shows that the gene is found in contig 52846 and it only has 1 exon between the positions 6829 and 6924 in the forward strand (+). The T-coffee shows a very bad alignment. So, we can discard also the second hit in this scaffold. Then, we can conclude that the predicted SelR2 is not found in the L. laticaudata genome. This might be due to the fact that the start positions of the scaffolds have predisposition to truncate.
In this case, 2 scaffolds showed significant hits when aligning with the SelR3 protein from human: BHFT01052183.1 (SelR3_laticauda_1) and BHFT01016323.1 (SelR3_laticauda_2). Due to their low query coverage we can not chose a good one. However, we believe that this two predictions correspond to the whole protein. In order to check our hypothesis, we performed a phylogenetic tree in which we can see that the combination of SelR3_laticauda_1 and SelR3_laticauda_2 correspond to the human SelR3. We can also see that the SelR2_laticauda is a copy of the human SelR3 due to their similarity.
The SelR3_laticauda_2 corresponds to the first part of the protein. Exonerate predicts 3 exons between 16832 and 30640 in the reverse strand (-). The T-coffe shows a good alignment, but, as we have just said only corresponds to the first part of the protein. The SelR3_laticauda_1 corresponds to the end part of the protein. Exonerate predicts 2 exons between 3919 and 6993 in the forward strand (+). The protein does not contain a Sec residue nor begins with a Met.
The T-coffe shows a good global alignment. Using Seblastian, a prediction is made but the positions do not correspond. This is due to the fact that the protein is compared to Protobothrops mucrosquamatus and not to human. No SECIS element is found, so, SelR3 is a cysteine-containing homolog, just as the human one.
Selenoprotein S (SelS) is a transmembrane selenocysteine-containing protein with roles in endoplasmic reticulum (ER) function and inflammation. It has been implicated in ER-associated protein degradation, and clinical studies revealed an association of its promoter polymorphism with cytokine levels and human diseases (Turanov AA et al., 2014).
In the SelS case, we got only one good prediction in the scaffold BHFT01004014.1 with high percentage of coverage and a low e-value. We conclude with Exonerate that the gene is found in contig 4013 and contains 5 exons between the positions 18927 and 27341 in the forward strand (+):

The T-coffee shows a very good alignment that covers the majority of the human protein and its value is around 1000. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Seblastian has not predict the alignment of the protein nor find a SECIS element.
So, the gene is found in the Laticauda laticaudata genome and shares homology with the human SelS, but we can not ensure that the predicted protein is a selenoprotein due to the absence of a SECIS element. Maybe, it has lost in our specie or the final part of the protein has changed so much that any prediction can not be done.
The Rdx family of selenoproteins includes mammalian selenoproteins SelH, SelT, SelV and SelW. These proteins possess a thioredoxin-like fold and a conserved CxxC or CxxU motif near the N terminus, suggesting a redox function (Dikiy A et al., 2007).
Selenoprotein H (SelH) is a 14-kDa mammalian nuclear protein involved in redox sensing and transcription (Novoselov SV et al., 2007)(Panee J et al., 2007).
In this case, we got only one good prediction in the scaffold BHFT01013888.1 with high query coverage percentage and low E-value. We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 13897 and it has only one exon that goes from the position 68152 to the 68232 in the forward strand (+):

The T-coffee showed a mostly good prediction but the alignment correspond to the final part of the protein. This prediction does not contains a selenocysteine residue nor a Met at the beginning of the protein. A better prediction is seen using Seblastian, the sequence begins with a Met and more aminoacids are shown in the N-terminal. One SECIS element is predicted in the 3’UTR end between positions 68816 and 68901 in the forward strand (+).
To conclude, this predicted protein is found in the L. laticaudata genome, but we can not know for sure if it is a selenoprotein due to the absence of the selenocysteine residue. This could be because the genome may be fragmented or because the region that contained the residue has been lost in L. laticaudata. Finally, it must be said that the human SelH and the predicted one are orthologous proteins.
Selenoprotein T (SelT) is an oxidoreductase localized to the Golgi complex and ER. It manifests a thioredoxin-like fold and it is involved in redox regulation and cell anchorage (Boukhzar L et al., 2016).
In the case of SelT, we got only one good prediction in the scaffold BHFT01014671.1 with a really high query coverage and low e-value. Exonerate shows us that the gene is found in contig 14688 with 5 exons predicted between the positions 34320 and 41681 in the reverse strand (-):

Using T-coffee, we can see that it is a very good alignment with a score around 1000. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Seblastian found a very similar predicted sequence and a SECIS element in the 3’UTR end between the positions 33643 and 33726 in the reverse strand (-).
We can conclude that the predicted SelT is found in the L. laticaudata genome and that it shares orthology with the human SelT. We can also say that the predicted protein is a selenoprotein, just as expected.
Selenoprotein V (SelV) is found only in placental mammals. It emerged by duplication from SelW. It is expressed exclusively in the testes, and therefore, it may be involved in reproduction, but its specific function is not known.
We have not found any relevant hit. So, the gene is not found in the Laticauda laticaudata genome. This might be due to the fact that it locates at the root of placental mammals.
Selenoprotein W (SelW) is the smallest selenoprotein containing a selenocysteine in a conserved - CXXU- motif. It contains both a selenocysteine residue as well as a bound glutathione molecule at residue Cys. SelW is expressed in various tissues, particularly in skeletal muscle and heart, and may have anti-oxidant activity which may be important in muscle growth (Jeon YH et al., 2014).
In this case, we got only one good prediction in the scaffold BHFT01019166.1 with high query coverage percentage and low e-value. Exonerate find that the gene is found in contig 19238 and it has 3 exons between the positions 23036 and 24652 in the reverse strand (-):

The T-coffee showed a value around 1000 and it also showed that it is a good prediction. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Seblastian find a better prediction that starts with a Met and has few more aminoacids in the N-terminal. Also, a SECIS element is present in the 3'UTR end between positions 22128 and 22214 in the reverse strand (-).
Taking all into account, we can conclude that the predicted SelW1 is found in the L. laticaudata genome and it is homologous to the human protein. Moreover, we can ensure that SelW1 is a selenoprotein, just as we expected.
With SelW2, we got only one prediction in the scaffold BHFT01058988.1 with low e-value and high query coverage percentage. Using Exonerate, we can know that the gene is in contig 59851 and contains 3 exons between the positions 12671 and 14402 in the forward strand (+):

The T-coffee shows a good alignment with value around 1000. This prediction does not contain a selenocysteine residue nor includes a Met at the beginning of the protein. No prediction nor SECIS elements could be predicted using Seblastian.
So, we can conclude that the SelW2 is found in the L. laticaudata genome and shares homology relationship with the human SelW2. However, we can not conclude that the predicted protein is a selenoprotein due to the absence of a Sec residue. Maybe in our specie the residue has lost and it is a cysteine homolog.
Selenoprotein U (SelU), a newly identified protein, is expressed highly in eukaryotes and possesses a conserved motif similar to that existing in other thiol-dependent redox regulating selenoproteins. In high mammalian species all SelU proteins exist in Cys form. Mammals contain three Cys-containing SelU proteins: SelU1, SelU2 and SelU3 (Sattar H et al., 2018).
In the SelU1 case, we got only one prediction in the scaffold BHFT01004697.1 with a 74% of coverage and a low e-value. We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 4696 and it has 5 exons between the positions 3867 and 17650 in the forward strand (+):

The T-coffee shows a very good alignment and a score value around 1000. This prediction contains a selenocysteine residue but does not include a Met at the beginning of the protein. Using Seblastian, we find a better prediction because the sequence begins with a Met. Also, a SECIS element is predicted in the 3’UTR end between the positions 17838 and 17902 in the forward strand (+).
Taken all together, we can conclude that the SelU1 is found in the Laticauda laticaudata genome and it is orthologous to the human SelU1. We can also ensure that it is a selenoprotein, not like the human SelU1, that is a cysteine-containing homolog. So, it has gained a new function as a selenoprotein.
In this case, we got only one prediction in the scaffold BHFT01039219.1 with high query coverage percentage and low e-value. Using Exonerate, we find that the gene is found in contig 39587 and 5 exons are predicted between the positions 38278 and 43505 in the reverse strand (-):

The T-coffee predicts a very good alignment and its score value is around 1000. This prediction does not contain a selenocysteine residue nor includes a Met at the beginning of the protein. No alignment nor SECIS elements can be predicted using Seblastian.
In conclusion, the predicted SelU2 is found in the L. laticaudata genome and it is orthologous to the human one. Moreover, we can ensure that it is not a selenoprotein due to the absence of the Sec residue and a SECIS element.
We have not found any relevant hit. So, the gene is not found in the Laticauda laticaudata genome.
The thioredoxin reductases (TrxRs) are a family of selenocysteine-containing oxidoreductases with mechanistic and sequence identity, including a conserved -CVNVGC- active site motif in its N-terminus, to glutathione reductases. The specificity of TrxRs is due to a second redox-active site motif in the C-terminus -GCUC- that is not found in glutathione reductase. TrxRs control the redox state of thioredoxins, key proteins involved in redox regulation of cellular processes. Mammals have three TrxRs isozymes (Ren X et al., 2017).
In the case of TR1, 2 scaffolds showed significant hits when aligning with the TR1 protein from human: BHFT01035671.1 (TR1_laticauda_1) and BHFT01015136.1 (TR1_laticauda_2). However, the one that showed a lower E-value and a higher query coverage was the one selected, BHFT01035671.1. To check which of the predictions was the most accurate and showed more similarity with the human TR1, we performed a phylogenetic tree. Due to the similarity of TrxRs family, another protein shows correspondence to TR1: the TR2_laticauda_3.
Using Exonerate, we find that the gene is found in contig 35979 and 12 exons are predicted between the positions 7161 and 18976 in the reverse strand (-):

The T-coffee shows a very good alignment with a score value around 1000. The predicted protein does not include a Sec residue nor include a Met at the beginning. Seblastian does not predict the protein nor a SECIS element.
So, we can conclude that the predicted TR1 is found in the L. laticaudata genome and that the human and the predicted proteins are orthologous. However, it is not a selenoprotein due to the absence of a Sec residue and a SECIS element, according to the Seblastian. Moreover, human TR1 has a Sec residue, so our specie has lost this residue.
In the case of TR2, 3 scaffolds showed significant hits when aligning with the TR2 protein from human: BHFT01035671.1 (TR2_laticauda_1), BHFT01015136.1 (TR2_laticauda_2) and BHFT01033478.1 (TR2_laticauda_3). We discard the first prediction due to the low similarity. In order to check which of the two predictions left is the most accurate and shows more similarity with the human TR2, we performed a phylogenetic tree. The one that shows lower E-value and higher query coverage is the one selected, BHFT01015136.1.
This prediction does not contain a selenocysteine residue nor includes a Met at the beginning of the protein. Exonerate shows that the gene is found in contig 15156 and 3 exons are predicted between the positions 15432 and 26802 in the forward strand (+):

The T-coffee shows a bad alignment, the major part of the protein is missing due to the fragmentation of L. laticaudata genome. The predicted protein does not include a Met at the beginning nor a Sec residue. Using Seblastian, a bad prediction is made because the positions do not correspond to the Exonerate ones because the protein is compared to Meleagris gallopavo.
To conclude, the predicted TR2 is found in L. laticaudata genome but it has diverged from the human one. We can not say that is a selenoprotein, but we can not ensure this because the human region containing the Sec residue might have lost in our specie.
In this case, we got only one prediction in the scaffold BHFT01035671.1 (TR3_laticauda) with high query coverage and low e-value. Looking at the phylogeny tree, there may be two possible explanations: the predicted TR3 has lost in our specie or that the branch tree is not sufficiently good and it is actually an orthologous to human TR3.
We performed the Exonerate, in case of the proteins were orthologous. We see that the gene is found in contig 35979 and it has 15 exons between the positions 6086 and 31145 in the reverse strand (-):
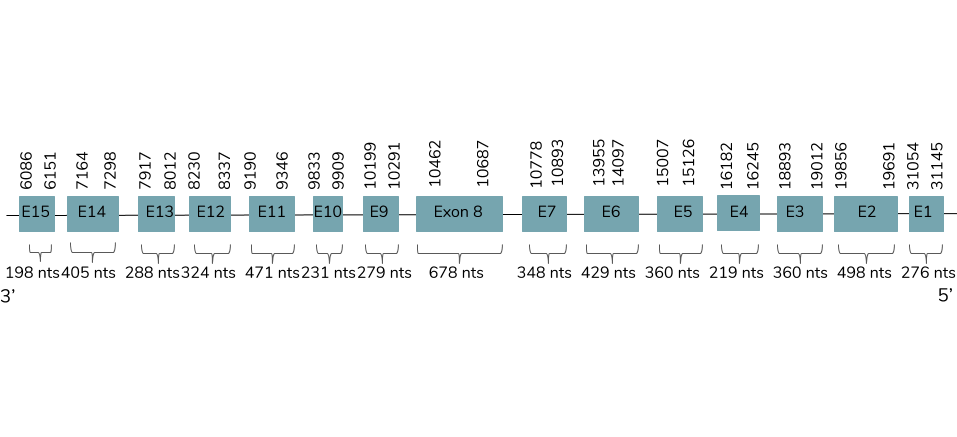
The T-coffee shows a good alignment and a score value around 1000. This predicted protein contains 3 selenocysteine residues but does not include a Met at the beginning. The human protein only has one Sec residue, so a gain in Sec residues has taken place. Seblastian does not predict the protein nor a SECIS element. So, it is not a selenoprotein.
In conclusion, the predicted TR3 is found in the L. laticaudata genome and shares a orthologous relationship with the human TR3.
The elongation factor eEFSec is a GTP-binding protein with similar affinity to GDP, thus it likely does not require a guanine nucleotide exchange factor. As for its tRNA-binding properties, eEFSec can only interact with Sec-tRNASec, but not serylated-tRNASec precursor or the canonical aminoacyl-tRNAs (Gonzalez-Flores JN et al., 2012). This translation factor is necessary for the incorporation of selenocysteine into proteins. It probably replaces EF-Tu for the insertion of selenocysteine directed by the UGA codon.
In this case, 4 scaffolds showed significant hits when aligning with the eEFSec protein from human: BHFT01045288.1, BHFT01045870.1, BHFT01031528.1 and BHFT01033925.1. Due to their low query coverage we can not chose a good one. However, if we base on the positions of the query start and end we observe that the protein might have been divided in 4 scaffolds. So, we chose the one with a higher query coverage: BHFT01045288.1.
Exonerate shows that the gene is found in contig 45778 with only one exon predicted between 27927 and 28595 in the reverse strand (-):
The predicted protein does not include a Sec residue nor a Met in the beginning. Also Seblastian does not predict the protein nor a SECIS element. So, the protein is not a selenoprotein. However, the fact that is divided in 4 scaffolds does not let us to confirm this.
Selenophosphate synthetase (SPS) is unique among the components of the Sec biosynthesis machinery in that it is often a selenoprotein itself. SPS catalyzes the synthesis of selenophosphate from selenide, ATP, and water, producing AMP and inorganic phosphate as products. Selenophosphate is the Se donor for the synthesis of Sec. SPS proteins are conserved from bacteria to human with approximately 30% identity and are found in all species known to encode selenoproteins. In vertebrats two paralogous SPS genes have been reported: SPS2, which is a selenoprotein, and SPS1 which is not and carries a Thr in vertebrates in place of Sec (Mariotti M et al., 2015).
In the case of SPS1, 2 scaffolds showed significant hits when aligning with the SPS1 protein from human: BHFT01001630.1 and BHFT01002322.1. However, the one that showed a lower E-value and a higher coverage was the one selected, BHFT01001630.1 (SPS1_laticauda_1). In order to check which of the predictions is the most accurate and shows more similarity with the human SPS1, we performed the following phylogenetic tree, in which we can observe SPS2 prediction due to their similarity.

We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 1629 and 8 exons are predicted. The gene predicted is located between the positions 15428 and 24655 in the forward strand (+):
The T-coffee shows a value of 1000 and it also shows that it is a very good alignment, so there has been almost no divergence. The predicted protein includes a Met at the beginning but no selenocysteine residue can be found. Also, Seblastian does not found any SECIS elements nor a protein prediction. So, the protein is not a selenoprotein, like the human one. Moreover, the predicted SPS1 is found in the L. laticaudata genome and presents orthology with the human SPS1.
For SPS2 protein, we find 2 scaffolds that showed significant hits when aligning with the SPS2 protein from human: BHFT01001630.1 and BHFT01002322.1. However, the one that shows a lower E-value and a higher coverage is the one selected, BHFT01002322.1 (SPS2_laticauda_2). In order to ensure that this is the most accurate prediction and the one that shows more similarity with the human SPS2, we performed the phylogenetic tree above in which we can observe SPS1 prediction due to their similarity.
We conclude with Exonerate that the gene is found in the Laticauda laticaudata genome in contig 2321 between the positions 67005 and 72421 in the reverse strand (-). In the gene 7 exons are predicted:

The T-coffee showed a value around 1000 and it also showed that it is a good prediction. The predicted protein does not include a Met at the beginning of the protein nor a Sec residue. The protein is predicted better with Seblastian because there are more aminoacids in the N-terminal and the prediction starts with a Met residue. Also, one SECIS element is found between 66580 and 66654 in the reverse strand (-).
So, although it has a SECIS element we can not say that it is a selenoprotein because of the inexistence of a Sec residue. Maybe the residue has been lost in our species changing its function or maybe the region of the human protein that contained the residue has changed in our species and it is not aligned. However, we can ensure that the predicted SPS2 protein is found in the L. laticaudata genome.
The SECIS binding protein 2 (SBP2) is an essential component of the machinery involved in co-translational insertion of selenocysteine into selenoproteins. As we have said in the introduction, Sec is encoded by the UGA codon, which normally signals translation termination. The recoding of UGA as Sec codon requires a SECIS element present in the 3' untranslated regions. SBP2 specifically binds to the SECIS element, which is stimulated by a Sec-specific translation elongation factor. Mutations in SBPS2 coding gene have been associated with reduction in enzymatic activity of DI2 and abnormal thyroid hormone metabolism (Hamajima T et al., 2012).
In this case, 3 scaffolds showed significant hits when aligning with the SBP2 protein from human: BHFT01003421.1 (SBP2_laticauda_1), BHFT01017391.1 (SBP2_laticauda_2) and BHFT01074757.1 (SBP2_laticauda_3). Due to their low query coverage we can not chose a good one. However, if we base on the positions of the query start and end we observe that the protein might have been divided. The first two scaffolds are a duplication and correspond to the end part of the protein (73% to 88% of the query), and the last one to the beginning (1% to 63%). So, we decide to study BHFT01003421.1 and BHFT01074757.1 scaffolds.
Using exonerate, we find that the first part of the protein (SBP2_laticauda_3) has one gene in contig 76186 and 5 exons are predicted between 13451 and 18916 in the forward strand (+). We also have performed Exonerate in the second part of the protein (SBP2_laticauda_1, the one with higher query coverage) and it shows that the gene is found in contig 3420 with 4 exons predicted between 84740 and 90360 in the reverse strand (-). This prediction does not contain any Sec residue nor includes a Met at the beginning of the protein.

No prediction of the protein nor any SECIS elements are predicted with Seblastian. So, we can conclude that the protein is truncated in two different scaffolds and that it is not a selenoprotein. Thus, the predicted SBP2 is found in the L. laticaudata genome.