INTRODUCTION
Selenium is a chemical element with symbol Se and atomic number 34. Selenium (Greek σελήνη selene meaning "Moon") was identified in 1817 by Jöns Jacob Berzelius, who noticed the analogy of this element to the previously known tellurium (named for the Earth) [5]. Selenium salts are toxic in high concentration, but trace amounts are necessary for cellular function in many organisms, including all animals. The National Institutes of Health set the Recommended Dietary Allowance for Selenium as 55 μg for both female and men [6]. Se is an essential component of several major metabolic pathways, including thyroid hormone metabolism, antioxidant defence systems, and immune function [7].
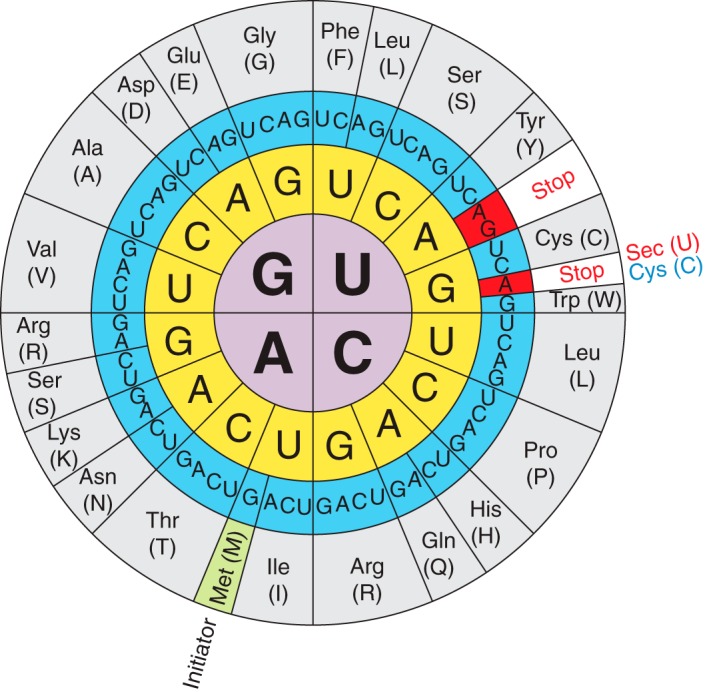
Figure 1. The genetic code illustrating the dual function of the UGA codon and that Sec is the 21st amino acid that is encoded by UGA [8].
We can find selenium in the cell as selenocysteine (Sec), the 21st proteinogenic amino acid. The molecular structure of selenocysteine is an analogue of the amino acid cysteine in which sulfur atom has been replaced by a selenium atom. It is encoded by the UGA codon, which is also the codon for the end of translation (Fig. 1) [8].
Selenocysteine is found in all lineages of life as an amino acid residue of selenoproteins, but not all organisms use them. In eukaryotes, selenoproteins show a mosaic occurrence, with some organisms, such as vertebrates and algae, having dozens of these proteins, while other organisms, such as higher plants and fungi, having lost all selenoproteins during evolution.
So far 45 selenoprotein families have been depicted in vertebrates. However, all these families together have never been certified to fuse into single specie. Besides, scientists have confirmed a decrease on its number during evolution process, especially after terrestrial settlement.
Most selenoproteins act as oxidoreductases that impede damage to cellular elements, repair this harm, control redox state of proteins, amongst other redox functions. These proteins play a major role in human health and disease so their deficiency cause some human diseases such as Keshan disease, Kashin-Beck disease, Myxedematous Endemic Cretinism and male infertility [9]
The common characteristic of selenoproteins is that they include minimum one residue of Sec, which is coded by UGA codon. The cis elements and trans factors are critical for Sec insertion during selenoprotein synthesis.
The basic trans-acting elements are:
- SPS2 (Selenophosphate synthetase 2)
- SecS (selenocysteine synthase)
- PSTK (phosphoseryl-tRNA kinase)
- eEFsec (eukaryoyic Sec tRNA-specific elongation factor)
- SBP2 (SECIS-binding protein 2)
- SeCys/tRNA[Ser]Sec.
All these factors are globally known as the selenoprotein translation machinery (Fig. 2).
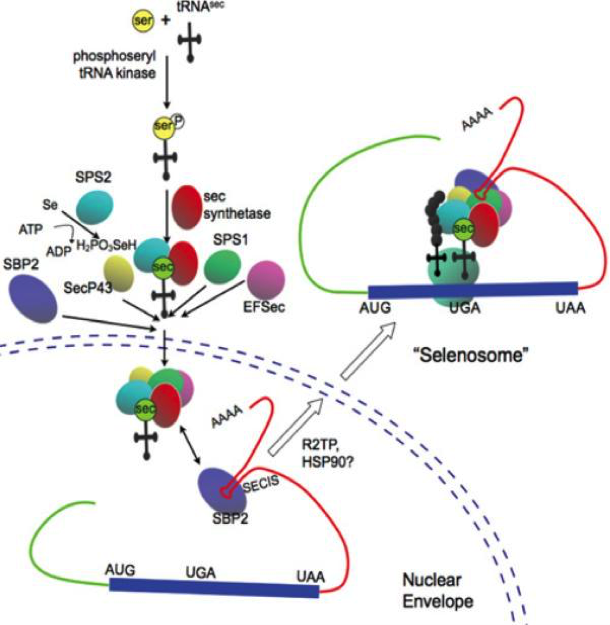
The basic cis-acting elements acting in the synthesis of a selenoprotein is the SECIS element (SElenoCysteine Insertion Sequence), an RNA stem-loop located in the 3′ untranslated region of selenoprotein genes that recruits several transacting factors to recode the UGA stop codon for Sec insertion [11]. SECIS are the binding site for SBP2 whose RNA-binding domain belongs to the L7Ae family of riboproteins (includes ribosomal proteins, snRNA and snoRNA-binding proteins).
To incorporate Sec into proteins, SBP2 must bind to SECIS element and EFsec binded to tRNASec must be gathered (Fig. 3). In addition, EFsec facilitates incorporation of Sec into the growing polypeptide. Finally to achieve the incorporation of the selenocysteine, the protein has to select the tRNA[Ser]Sec, which is taken to the UGA codon [12].
These factors assembly on selenoprotein mRNA comes before the translation of UGA as Sec. Oxidation of SBP2 leads to its import into the nucleus, where its reduction by nuclear-specific GPX and TRXR proteins is followed by CRM-1-dependent nuclear export. Supplementary cofactors might contribute to selenoprotein synthesis. The requirement of diverse specialized factors dedicated to incorporation of Sec into proteins suggests the evolutionary significance of selenoproteins [10].
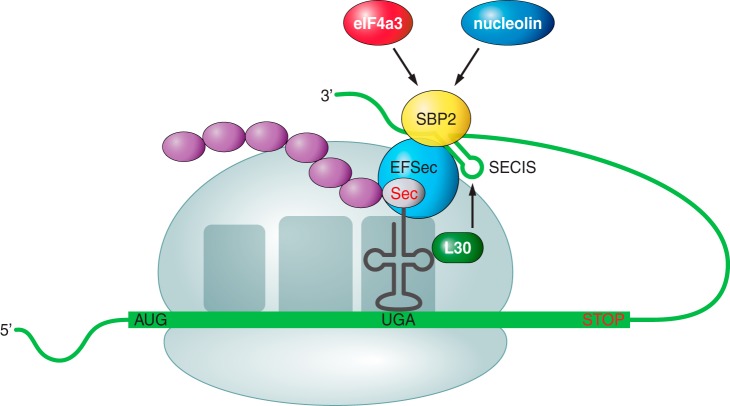
The tRNA for Sec (tRNASec) identifies the codon UGA, which functions in many mRNAs as a stop codon [13]. Thus the efficiency of UGA-selenocysteine recoding is the limiting stage of selenoprotein synthesis [14]
Sec is the only aminoacid whose biosynthesis is produced in its own tRNA called Sec tRNA[Ser]Sec. The tRNA[Ser]Sec is initially made with serine in a reaction catalyzed by seryl-tRNA synthesase (SerS) to make seryl-tRNA[Ser]Sec that provides the skeleton for Sec biosynthesis.
Serine is conjugated to tRNASec by SerS and then PSTK phosphorylates the Ser (converting it into phosphoserine) using a molecule of ATP. Following this, SPS 2 prepares the Selenium to be incorporated and SecS binds the atom to the serine, completing the synthesis of the tRNA. This tRNA is not recognized by usual elongation factors, and is instead bound to the specific factor eEFSec [13]. Figure 4 illustrates this process.
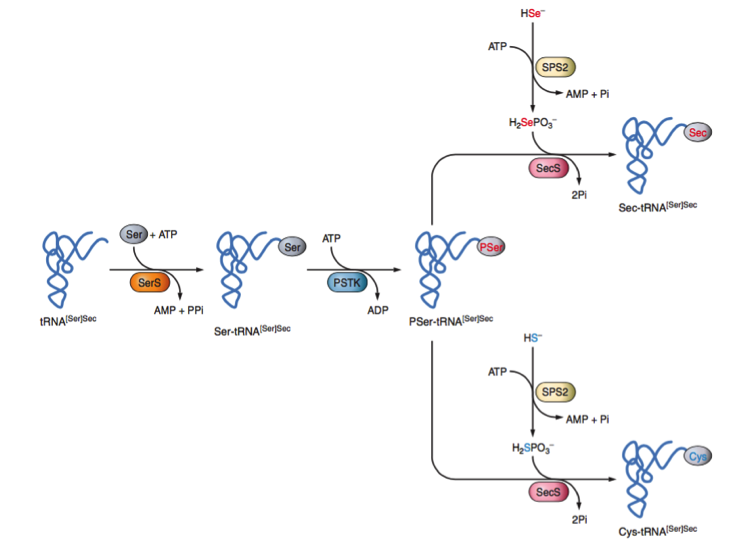
and the Sec machinery-based pathway for synthesis of Cys [14]
We can distinguish two major groups of mammalian selenoproteins depending on their function. The first group is constituted by housekeeping genes and the second group includes stress activated proteins. The housekeeping genes are less influenced by the amount of selenium consumed in the diet and participate in critical functions for cellular survival. On the other hand, stress-related selenoproteins are not indispensable the survival and are less expressed in a situation of selenium deficit [16]. A brief description of the several selenoproteins families is summarized in the following table:
| Name | |
| Glutathione peroxidase is a family of 8 proteins with peroxidase activity that show strong antioxidant activity and protect cell membranes and other cellular components against oxidative damage |
|
| Thioredoxin reductase is a selenocysteine containing enzyme which catalyses the NADPH dependent reduction of thioredoxin and therefore plays a regulatory role in its metabolic activity. There are three described in mammals: TR1, TR2 and TR3 |
|
| Iodothyronine deiodinase enzymes are a family of 3 enzymes related to thyroid hormones because they catalysed activation or inactivation reactions. DI1 and DI2 have an important role in the production of T3 from T4. DI3 converts T4 to T3 reverse |
|
| Sel15 is a thioredoxin-like protein that may have a redox function and may be involved in the control of glycoprotein folding process |
|
| Selenoprotein H (Sel H) is a thioredoxin fold-like protein that contains a conserved Cys-X-X-Sec motif (X is any amino acid). Its expression is widely distributed throughout a variety of tissues and relatively high in early stages of embryonic development |
|
Selenoprotein I (also called hEPT1) is expressed in all the tissues in which phospholipid synthesis takes place so it is expressed ubiquitously |
|
| Selenoprotein K works as an antioxidant because it decreases the levels of intracellular ROS and protects cardiomyocytes from oxidative stress |
|
| Selenoprotein M is involved in redox reactions. It is interesting to know that the gene responsible for the onset of Alzheimer inhibits SelM |
|
Selenoprotein N regulates calcium mobilization required for normal muscle development and differentiation. Mutations in its gene cause some kinds of myopathies |
|
Selenoprotein O has an unknown localization, distribution and function. However, it has a domain that suggest that its function it's related to redox reactions |
|
| Selenoprotein P has multiple selenocysteine residues and this suggests a role in selenium transport to different tissues |
|
| Selenoprotein R (also called Methionine sulfoxide reductase B or MsrB) is a member of a family of proteins widely distributed throughout the tissues. It catalyses the reduction of oxidized methionine residues obtained in response to increase in ROS elements |
|
| Selenoprotein S is a transmembrane protein that has been suggested to participate in the removal of misfolded proteins from the ER lumen for degradation, to protect cells from oxidative damage and ER stress-induced apoptosis. It is related to type 2 diabetes, cardiovascular disease and inflammation and has an important role in controlling inflammatory response |
|
| Selenoprotein T is believed to act in cellular stress and in the activation of adenylate cyclase activator polypeptide in the pituitary gland by regulating calcium |
|
| Selenoprotein V expression is restricted to testis but its function is still unknown. However, the sequence of the SelV is homologous with other selenoproteins with folding as thioredoxin, so it is believed to be involved in redox functions |
|
| Selenoprotein W is a small selenoprotein highly conserved in mammals that is expressed depending on the amount of selenium in the body. As it binds glutathione with a high affinity, it is thought that its function is to act as an antioxidant |
|
Methionine Sulfoxide Reductase A is an enzyme that catalyses the reaction from methionine sulfoxide to methionine. This process also forms part of oxidative stress mechanisms. |
| Name | |
Eukaryotic elongation factor (eEFSec). It is involved in selenocysteine incorporation. |
|
Phosphoseryl-tRNA kinase is found to be highly conserved in evolution, hence suggesting that it plays an important role in selenoprotein biosynthesis and/or regulation. Catalyses the reaction from seryl-tRNA Ser (Sec) to the phosphoryl form using a molecule of ATP. |
|
SECIS-binding protein 2 is a nuclear protein that functions as a SECIS binding protein and recruits eEFSec and other factors in order to recode the UGA codon. |
|
tRNA Sec 1 associated protein 1 regulates selenoprotein expression forming a complex with tRNASec. |
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (also called LP; SLA; PCH2D; SLA/LP). The amino acid selenocysteine is the only amino acid that does not have its own tRNA synthetase. Instead, this amino acid is synthesized on its cognate tRNA in a three step process. The protein encoded by this gene catalyzes the third step in the process, the conversion of O-phosphoseryl-tRNA(Sec) to selenocysteinyl-tRNA(Sec). |
|
Selenophosphate synthetase (SPS) are selenoproteins essentials for selenoprotein synthesis. There are two isoforms in mammals: SPS1 and SPS2. Selenophosphate 1 is a cysteine homologue, whereas selenophosphate synthetase 2 (SPS2) is a selenoprotein which plays a role in the synthesis of the Sec aminoacid. |
West Indian manatees (Trichechus manatus), most commonly known as "big cows", are big, slow-moving, gentle vegetarians. They are the largest surviving member of the aquatic mammal order Sirenia (which also includes the dugong and the extinct Steller's sea cow) [17]. The West Indian manatee is divided into two subspecies, the Florida manatee (T. m. latirostris) and the Antillean or Caribbean manatee (T. m. manatus) [18].
The Florida manatee is a long-lived marine mammal, dark gray in color, and averaging about 3 m in length and between 363 to 544 kg in weight [17]. It exhibits a wide range of morphological adaptations for its tropical, shallow-water, herbivorous life. Among these adaptations are an elongated streamlined body, enlarged rostrum, low-crowned rounded molars with horizontal replacement, elongated diaphragm with orientation in the frontal (dorsal) plane, fore flippers, lack of hind legs, a caudal fluke, and pronounced skeletal pachyosteosclerosis—a hydrostatic aid [19].
The Florida manatee subspecies is listed as Endangered on the basis of a population size of less than 2,500 mature individuals and the population is estimated to decline by at least 20% over the next two generations (estimated at ~40 years) due to anticipated future changes in warm-water habitat and threats from increasing watercraft traffic over the next several decades [20].
Here we show a table with the taxonomic classification [21] :
| Kingdom | Animalia |
| Phylum | Chordata |
| Subphylum | Vertebrata |
| Superclass | Gnathostomata |
| Class | Mammalia |
| Subclass | Theria |
| Order | Sirenia |
| Family | Trichechidae |
| Genus | Trichechus |
| Species | Trichechus manatus |
| Subspecies | Trichechus manatus latirostris |
The elephant (Loxodonta africana) was chosen as the reference genome as it is the closest one phylogenically from the available species in SelenoDB. They share the same superorder: Afrotheria. It is a superorder of placental mammals comprising three clades: Afroinsectiphilia, Pseudungulata and Paenungulata, where the latter includes Trichechus manatus latirostris and Loxodonta africana [22].