We executed a computational analysis in order to compare Annarrhichtys ocellatus’s selenoproteome with the reference zebrafish selenoproteins and machinery. The search was supplemented with the analysis of SECIS elements via Seblastian, and with the phylogenetic analysis of proteins belonging to the same family. The resulted orthologous proteins are presented one by one, together with their gene representation.
According to the results we obtained, not all selenoproteins described in zebrafish are present in Anarrhichtys ocellatus. Consequently, our species of interest has gained and lost some of the Sec residues found in zebrafish.
The reasoning of our results is discussed hereunder.
Sel15 is thioredoxin-like fold protein that localizes to the endoplasmic reituculum (ER). Its main function is the regulation of the redox homeostasis in the ER. It also may be involved in the quality control of protein folding. In zebrafish, Sel15 is a selenoprotein that contains a selenocysteine [1].
We found that Sel15 is conserved in Anarrhichtys ocellatus in the scaffold ML171018.1, which showed good E-value and identity at TBLASTN. The T-coffee showed also a good score for the alignment of sequences and both Sec residues aligned together. The predicted protein had the exact same length. Apart from this, Seblastian using the fastasubseq sequence predicted a known selenoprotein (Sel15), and found one SECIS element.
Considering these results, we were able to determine that Anarrhichtys ocellatus has a Sec-containing Sel15 located in the forward strand of the scaffold. Its encoding gene has 5 exons. Te mRNA has a grade A SECIS element at the 3’UTR region in the same strand.
SELENOE is protein found in the ER whose function is not known. It is only found in fish, so in zebrafish, SELENOE is a selenoprotein [1].
We have also found SELENOE in Anarrhichtys ocellatus, concretely in the scaffold ML171012.1. TBLASTN showed great identity and T-coffe score was high (994).
Exonerate showed that our predicted nucleotide sequence had 5 exons and followed a forward direction. The amino acid sequence also contained the same selenocysteine present in Zebrafish. Seblastian predicted for this protein one SECIS element in the same strand and close to the coding sequence.
Therefore, we could state that SELENOE is conserved as a selenoprotein in Anarrhichtys ocellatus.
This family of proteins is widespread in all three domains of life: eukarya, archea and bacteria. GPxs play a large number of physiological functions involved in hydrogen peroxide signaling, detoxification of hydroperoxides and maintaining cellular redox homeostasis. This family is composed by several paralogs very similar to each other, that is why when aligning each member of the family, many scaffolds appeared to be probable. We tried to choose different scaffolds whenever it was been possible. In zebrafish, Gpx1, Gpx2, Gpx3 and Gpx2 are selenoproteins, whereas GPx7 and GPx8 are cysteine homologues [1].
GPx1 is the most abundant selenoprotein in mammals. It is a cytosolic enzyme that catalyzes gluthatione (GSH)-dependent reduction of hydrogen peroxide to water. This enzyme is expressed in all cell types. Its main function is the regulation of metabolic processes.
For GPx1a, TBLASTN showed 9 possible scaffolds that contained hits. The scaffold ML171004.1 showed a lower E-value and higher identity when compared to the zebrafish sequence. Moreover, the phylogenetic analysis supported that decision, as this scaffold was more closely related to the ancestral protein. Exonerate indicated that the gene that encodes for GPx1a has 2 exons in the reverse strand. Seblastian predicted a multiple SECIS elements but only one was acceptable because it was the only one located at the 3’UTR region of the exons.
In summation, in Anarrhichtys ocellatus GPx1a is a selenoprotein found in the reverse strand of the scaffold ML171004.1. Its gene has 2 exons and its mRNA has a SECIS element at the 3’UTR region.
TBLASTN for GPx1b also proposed 9 possiblee scaffolds. ML171004.1 and another candidate (ML171007.1.) showed the best identity score and the lower E-value. Even though ML171007.1 was more closely related phylogenetically, the quality of T-Coffee was way better for ML171004.1, so that was the accepted scaffold. Seblastian predicted a SECIS element at the 3’UTR region.
When analyzing both GPx1a and GPx1b protein predictions together we noticed that they were almost the same sequence. There was a little variation at the beginning and the end of the sequences. However, this variation could be due to problems in genome annotation. Also, when we compared the positions of the exons in the gene, we observed that the positions were almost identical and in the same scaffold.
Taking all that into consideration, we suggest that in Anarrhichtys ocellatus GPx1a and GPx1b are the same protein, found in the same scaffold (ML171004.1) and with the same structure.
GPx2 is the gluthatione peroxidase found in the epithelium of the gastrointestinal tract. It plays a role in the development of cancer but that is still unknown.
In Anarrhichtys ocellatus TBLASTN showed 9 possible scaffolds, with ML171010.1 being the one with the lowest E-value and the higher score. The sequence predicted with this scaffold also appeared to be evolutively closer to zebrafish reference nucleotide sequence in the phylogenetic tree. Our predicted sequence has 2 exons in the forward and the selenocysteine residue is conserved. Seblastian predicted a SECIS element at the 3’UTR region.
In Anarrhichtys ocellatus, GPx2 is a selenoprotein found in the scaffold ML171010.1 and its mRNA contains a SECIS element.
GPx3 is secreted from the kidney and it is the major GPx form in plasma.
In our studied species GPx3a can be found in the scaffold SAUM01028039.1. TBLASTN did not show the best results for this scaffold, but the T-Coffee was able to successfully align the predicted protein with the reference zebrafish one. The Sec residues in both species aligned together. To back up this decision, phylogeny indicated that the predicted protein from SAUM01028039.1 was more closely related to GPx3 in zebrafish. Seblastian predicted a SECIS element at the 3’UTR region. In Anarrhichtys ocellatus, GPx3a is conserved as a selenoprotein found scaffold SAUM01028039.1. Its gene contains 4 exons in the reverse strand. In the mRNA there is a SECIS element at the 3’UTR region.
GPx3b was found in scaffold ML171010.1 from Anarrhichtys ocellatus genome. The scaffold was also selected between 9 candidates since it showed a clear relationship in the phylogenetic tree and had a lower E-value and higher identity score. The predicted protein sequence has 2 exons in forward strand. T-Coffee alignment showed a high score and both Sec residues coincided. Seblastian predicted a SECIS element at the 3’UTR region. Therefore, GPx3b is conserved as a selenoprotein in Anarrhichtys ocellatus.
It is common to find gene duplications in fish, as their whole genome duplicated early in their evolution. Several proteins have been found duplicated in fish. It has been demonstrated, that two of those duplicated selenoproteins are GPx1 and GPx3, giving as a result GPx1b and GPx3b respectively. As we have mentioned before, there is only one GPx1 in our species of interest, but GPx3 has a duplication (GPx3b) in another scaffold.[11,12,13]
Gpx4 is a monomeric protein expressed in a wide range of cell types and tissues. GPx4 is involved in the reduction of membrane-bound phospholipids and inhibition of lipid peroxidation and decomposition. This enzyme is also implicated in regulation protein tyrosine phosphatases (PTPs).
GPx4a is present in ML171018.1 scaffold from Anarrhichtys ocellatus genome. It is the best scaffold from a total of 9 candidates considering TBLASTN E-value and score, as well as phylogeny. Exonerate showed the presence of 3 exons in the forward strand of its gene. Seblastian predicted a grade A SECIS element in the 3’UTR region.
GPx4b is located in the scaffold ML171018.1. It starts at the position 6,110,058 and finishes at 6,112,094 of the forward strand. Its gene has 4 exons and its structure is depicted below. Seblastian was able to predict one known selenoprotein and one SECIS element using the nucleotide sequence predicted in Anarrhichtys ocellatus’ genome. The SECIS element is located in the positions 6,112,456 to 6,112,535 in the forward strand..
As we can observe both GPx4 were found in the same scaffold at the same positions. Both proteins were encoded by a gene with 4 exons in the forward strand. They both had the same predicted SECIS element and phylogenetically, they branched together. Just like GPx1, all data point to the fact that Anarrhichtys ocellatus has one type of GPx4 as both proteins predicted are the same protein.
GPx7 and GPx8 are both gluthathione peroxidases and two important antioxidant enzymes. Unlike other proteins in the family, their mRNAs do not contain the UGA codon in zebrafish, thus they cannot code for selenoprotein.
GPx7 is in the scaffold ML171105.1 of our studied fish. TBLASTN and the phylogenetic study supported the fact that ML171105.1 was the best scaffold to analyze. Exonerate showed the presence of 4 exons in the forward strand of the gene. The T-Coffee showed good alignment and neither of the sequences contained selenocysteine. Seblastian predicted a SECIS element, but it was rejected due to the large distance that separated it from the protein sequence. Therefore, we could conclude that, in Anarrhichtys ocellatus, GPx7 is conserved as a cysteine homologue.
The last GPx protein is GPx8. GPx8 is located in the scaffold ML171018. ML171018 showed strong evolutionary relationship with the reference protein in zebrafish. Exonerated indicated the presence of 2 exons in the forward strand of the gene. T-Coffee successfully aligned both zebrafish and Anarrhichtys ocellatus GPx8 sequences. Interestingly, T-Coffee revealed a Sec residue incorporation in our species. These results correlated with the finding of a viable SECIS element in 3’UTR region of the gene. Thus, we were able to describe that GPx8 became a selenoprotein in Anarrhichtys ocellatus, and its correct synthesis depended on the presence of the SECIS element.
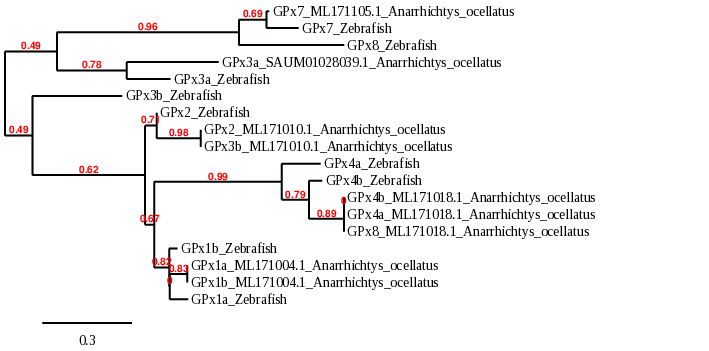
When looking at the whole family phylogeny, we can corroborate that they are very similar to each other. As GPx1a and GPx1b appear together we can hold our hypothesis, that is, they are the same protein. It also happens with GPx4a and GPx4b. Contrarily, GPx3a and GPx3b have significant differences, as they diverged before. In general, we can see a clear correspondence with Zebrafish homologous proteins, except in the case of GPx8.
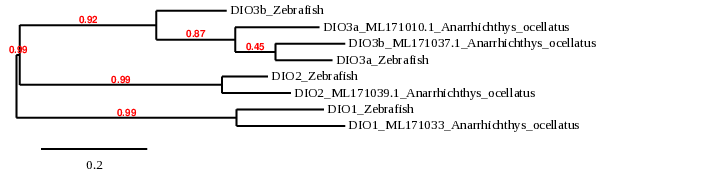
This family consists of three paralogous selenocysteine-containing proteins (DIO1, DIO2 and DIO3) that regulate thyroid hormone activity by reductive deiodination. They all have different localization and tissue expression [14]. In this case, Anarrhichtys ocellatus presents a duplication for DIO3, which is also expected to be duplicated in all bony fishes as a product of their whole-genome duplication. In zebrafish, all DIO are selenoproteins[9].
DIO1 and DIO2 enzymes are both located on the plasma membrane. They convert inactive form of thyroid hormone (T4) into the active form (T3), by outer ring deiodination [14].
When analyzing our species, DIO1 was found in scaffold ML171033.1. This scaffold showed the lowest E-value and the highest identity in TBLASTN results. The predicted protein contained 3 exons in the reverse strand. The alignment with the reference protein in T-coffee was very reliable with a score of 999. Selenocysteine residue was found conserved. Seblastian predicted 4 SECIS but only the one fitted our requisites (same strand and close position at 3’UTR). Therefore, Anarrhichtys ocellatus contains DIO1 as a selenoprotein.
DIO2 was found in scaffold ML171039.1 from Anarrhichtys ocellatus genome. This scaffold was chosen by T-Coffee best results and phylogenetic proximity. The predicted protein also had 3 exons in the reverse strand and the selenocysteine residue was conserved (as shown in T-Coffee alignment). A SECIS element was predicted according to the selenocysteine finding. These findings indicated the presence of DIO2 as a selenoprotein in Anarrhichtys ocellatus’ proteome.
Generally, the protein DIO3 is localized in the endoplasmic reticulum (ER), and inactivates both T4 and T3 [14].
For DIO3a we found four possible scaffolds. Scaffolds ML171010.1 and ML171037.1 showed similar results in TBLASTN and T-coffee, in which both scaffolds showed a good alignment and the presence of the Sec residue in the same position as zebrafish. However, when we analyzed the phylogenetics, we found that ML171010.1 was more closely related to the same protein in zebrafish, so we chose that scaffold. The predicted protein was 15 amino acids shorter than the one extracted from zebrafish, possibly due to a misprediction from exonerate. Seblastian predicted the selenoprotein DIO3a and a grade A SECIS element in the 3’UTR region.
Considering these results, we were able to determine that DIO3a is a Sec-containing protein in Anarrhichtys ocellatus, located in the forward strand of the scaffold ML171010.1. Its encoding gene has one grade A SECIS element in the same strand, at the 3’UTR of its only exon.
Similarly to DIO3a, for DIO3b, four scaffolds showed significant hits when aligning the corresponding protein from Zebrafish to Anarrhichtys ocellatus’ genome. The scaffolds ML171010.1 and ML171037.1 showed lower E-value and higher identity than the other candidates. Although the results from TBLASTN, T-Coffee and phylogeny were very similar between both of them, the scaffold ML171037.1 was the one we chose, because it had better alignment. Moreover, it had a selenocysteine at the same location as the zebrafish. The predicted protein had almost the same size as the one found in zebrafish; two amino acids were added and only the last one was missing. Seblastian predicted the selenoprotein DIO3b and a grade A SECIS element in the 3’UTR region.
Taking these results into account, we were able to determine that DIO3b is a Sec-containing protein in Anarrhichtys ocellatus, located in the forward strand of the scaffold ML171037.1. Its encoding gene has two exons and a grade A SECIS element at the 3’UTR, in the same strand.
MsrA catalyzes the repair of the S enantiomer of oxidized methionine residues in proteins[14]. In zebrafish, MsrA are not selenoproteins, they are cysteine homologues[9].
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171010.1 showed lower E-value and greater identity at TBLASTN than the other candidate (ML171028). The predicted protein had missed the first 17 amino acids, but it shows great similarity. It could be the result of an uncompleted sequence prediction. Like its zebrafish homologue, the predicted protein did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
In summary, in Anarrhictys ocellatus, MsrA is a cys-containing homologue protein related to Sec production. Its gene is located in the scaffold ML171010.1 and contains 6 exons in the reverse strand.
SELENOH is a protein that localizes in the nucleoli. It has gluthatione peroxidase activity and it is involved in the regulation of transcription of genes related to gluthatione synthesis and detoxification of enzymes [1]. In zebrafish, SELENOH exhibits a tumor-supressor function, consistent with its role in regulating redox homeostasis, inflammation and DNA damage during embryonic development. SELENOH deficiency collaborates with a loss of p53 function and also inducing inflammatory genes [18]. In zebrafish, SELENOH is a selenoprotein.
When analyzing Anarrhichtys ocellatus, we found one possible scaffold. The scaffold ML171030.1 showed great e-value and identity at TBLASTN. The t-coffee also showed a good score for the alignment of sequences and both Sec residues aligned together. The predicted protein missed the first 4 amino acids, so it do not contain the amino acid Methionine. Apart from this, using Seblastian with the fastasubseq sequence predicted a known selenoprotein (SelH) and a grade A SECIS element at the 3’UTR region.
As a result, in Anarrhichtys ocellatus, SELENOH is a selenoprotein, its gene is located in the reverse strand and it has 3 exons. The SECIS element is found downstream in the same strand.
SELENOI is a transmembrane protein only found in vertebrates[1], and its physiological function have been well described in mammals, where it is expressed in many tissues, particularly high in the brain, placenta, liver and pancreas [19]. It has been recently ascribed to have a role in neural development and maintenance of plasmalogen in humans [20]. In zebrafish, SELENOI is a selenoprotein.
When analyzing Anarrhichtys ocellatus, we found four possible scaffolds. In contrast to what we expected, any of these had the Sec residue and the alignments were very poor in all cases because of a significant loss of amino acids, and none of them aligned with the Sec residue of zebrafish. We finally choosed the scaffold ML171024.1 being the best alignment obtained in the t-coffee even though we cannot consider this scaffold as significant. Accordingly, Seblastian using the fastasubseq sequence was not able to predict any selenoprotein nor SECIS elements in any of the scaffolds.
Taking this data into account, in Anarrhichtys ocellatus, it is probably that SELENOI had been lost or that the sequence had been missanotated, which leads to a poor prediction of the protein.
SELENOJ is today found only in fishes, among vertebrates [5]. In contrast to all known eukaryotic selenoproteins, it does not exist in mammalians, not even a Cys homologue.
In zebrafish, SELENOJ has a major and homogeneous expression in the eye lens in early stages of development [21]. Contrarily to the majority of selenoproteins that have enzymatic functions, data suggest that SELENOJ could have a structural role. Data suggest that it may have been derived from ancestral ADP-ribosylation enzymes [21]. However, the function of SELENOJ is not fully understood.
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171018.1 showed a better e-value and identity at TBLASTN. The t-coffee showed also a better score for the alignment of sequences and both Sec residues aligned together. Moreover, the phylogeny study indicated that the sequence found in that scaffold was significantly closer related to the zebrafish SELENOJ [Fig. phylogeny]. The predicted protein was 32 amino acids shorter at the end, but it showed great similarity. It coincides at the start, with the first amino acid being a Methionine.
Seblastian using the fastasubseq sequence predicted one known selenoprotein (SelJ) and a grade A SECIS element at the 3’UTR region.
Consequently, in Anarrhichtys ocellatus, SELENOJ1 is a selenoprotein, its gene is located in the forward strand and it has 8 exons. The SECIS element is found downstream in the same strand.
This protein is a transmembrane protein located in the ER. Like SELENOS, Its functions involve: the ER-associated degradation of misfolded proteins, the reduction of disulfides in glycoprotein substrates and the mediation of anti-inflammatory effects of Se. In zebrafish, SELENOK is a selenoprotein.[1]
When analyzing Anarrhichtys ocellatus, we found one possible scaffold. The scaffold ML171008.1 showed good e-value and identity at TBLASTN. The t-coffee showed also a good score for the alignment of sequences and both Sec residues aligned together. The predicted protein had exactly the same length. Apart from this, Seblastian using the fastasubseq sequence was not able to predict the selenoprotein, but it found a grade A SECIS element at the 3’UTR region. Thus, we speculate that because it has a Sec residue it is, in fact, a selenoprotein (SelK).
Taking this into account, in Anarrhichtys ocellatus, SELENOK is a selenoprotein, its gene is located in the reverse strand and it has 4 exons. The SECIS element is found downstream in the same strand.
SELENOL is today found only in fishes, among vertebrates. This selenoprotein, characteristically with SELENOP, has multiple Sec residues. The two Sec residues in SELENOL are inserted with the help of a single SECIS element[5].
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171028.1 showed a better e-value and identity at TBLASTN. The t-coffee showed also a better score for the alignment of sequences and the two Sec residues aligned with the two Sec residues of zebrafish. Moreover, the phylogeny study indicated that the sequence found in that scaffold was significantly closer related to the zebrafish SELENOL [Fig. phylogeny]. The predicted protein had missed the first 135 amino acids, but it shows great similarity. It could be an uncompleted prediction of the sequence.
Seblastian using the fastasubseq sequence predicted one known selenoprotein (SelL) and a grade A SECIS element at the 3’UTR region.
As a result, in Anarrhichtys ocellatus, SELENOL is a selenoprotein, its gene is located in the forward strand and it has 4 exons. The SECIS element is found downstream in the same strand.
SELENOM is highly conserved from plants to humans and localizes to the endoplasmic reticulum. SELENOM is thioredoxin-like fold protein that participates in the formation of disulfide bonds and can be implicated in calcium responses. It is highly expressed in the brain, where it is believed to have a role in neuroprotection. SELENOM is also associated with the catalysis of free radicals and has been associated with Alzheimer’s disease (AD) [1,22]. However, studies in fishes remain limited. In zebrafish, SELENOM is a selenoprotein.
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171020.1 showed a better e-value and identity at TBLASTN. The t-coffee also showed a better score for the alignment of sequences. However, none of the scaffolds incorporate a Sec residue. The t-coffee sequence alignment did not associate the Sec residue of zebrafish with any amino acid, because the predicted sequence of Anarrhichtys ocellatus was too short. The predicted protein was missing the first 41 amino acids, where the zebrafish sequence contained the Sec residue.
Seblastian using the fastasubseq sequence predicted one known selenoprotein (SelM) and a grade A SECIS element at the 3’UTR region. If we consider that our scaffold sequence did not contain a Sec residue but it still contains a SECIS element, one possible explanation is that SELENOM could have been a selenoprotein that no longer contains the Sec residue, although we cannot confirm which amino acid replace the position of this residue. On account of the fact that it still contains the SECIS element, we believe that it has been a relatively evolutionary recent lost of the selenoprotein.
Analyzing Anarrhichtys ocellatus, what we obtained is a gene located in the reverse strand that has 3 exons. The SECIS element is found downstream in the same strand.
SELENON is an ER-resident transmembrane glycoprotein expressed during embryonic development and in adult tissue. This protein is required for early muscle development and differentiation. In zebrafish, SELENON is a selenoprotein[1].
When analyzing Anarrhichtys ocellatus, we found one possible scaffold. The scaffold ML171010.1 showed good e-value and identity at TBLASTN. The t-coffee also showed a good score for the alignment of sequences and both Sec residues aligned together. The predicted protein had practically the same length, except for the last 2 amino acids. Moreover, using Seblastian with the fastasubseq sequence predicted a known selenoprotein (SelN) and a grade A SECIS element at the 3’UTR region.
As a result, in Anarrhichtys ocellatus, SELENON is a selenoprotein, its gene is located in the reverse strand and it has 12 exons. The SECIS element is found downstream in the same strand.
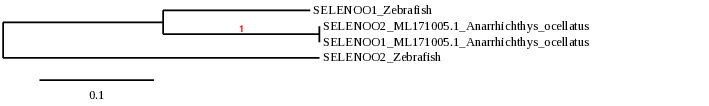
SELENOO is a mitochondrial Se-containing protein that is not well characterized yet, so its function is still unclear. Ultimately, a redox-active role has been suggested [23]. The latest research reveals an essential role of SELENOO in cartilage function through regulation of chondrocyte proliferation and apoptosis in mammalians, although the mechanism is still unknown [24]. Therefore, this protein is induced during chondrocyte differentiation and shows crucial roles in cartilage formation.
In zebrafish, SELENOO proteins are selenoproteins and contain Sec residues and it has also been described additional copies of SELENOO, probably due to the whole genome duplication in the early evolution of fishes[5]. Zebrafish has SELENOO1 and the duplicated SELENOO2 protein.
Nonetheless, we found that Anarrhichtys ocellatus’ genome, there is only one copy of SELENOO. When analyzing SELENOO1 and SELENOO2 in our species of study, we observed that they were both the same protein.
The two proteins presented two possible scaffolds. The scaffold ML171005.1 showed a better e-value and identity at TBLASTN. The t-coffee also showed a better score for the alignment of sequences and both Sec residues aligned together. Even though the phylogeny study indicated that the sequence found in that scaffold was not the most significantly closer related to the zebrafish’s SELENOO protein, we choosed the one with a better alignment in the t-coffee. The predicted protein was practically the same as the reference except for 3 amino acids at the middle of the sequence.
Seblastian using the fastasubseq sequence predicted one known selenoprotein (SelO) and a grade A SECIS element at the 3’UTR region. The SECIS found for SELENOO1 and SELENOO2 was the same element.
As a result, we can conclude that the predicted protein SELENOO2 refers to the same protein. Thus, we suggest that in Anarrhichtys ocellatus, the SELENOO family consists in only one protein found in scaffold ML171005.1. Its gene is located in the reverse strand and it has 9 exons. The SECIS element is found downstream in the same strand.
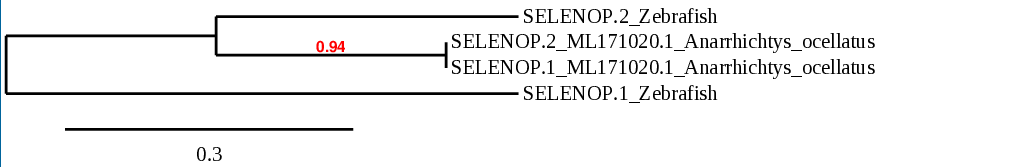
SELENOP is a secreted selenoprotein synthesized predominantly in the liver, so it accounts for almost 50% of the total Se in plasma. It contains more than one selenocysteine residue. SELENOP’s main function is the transportation of Se to peripheral tissues and preserving the function of said tissued under conditions of limiting Se. In zebrafish, SELENOP is a selenoprotein[27].
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171020.1 showed better E-value and identity at TBLASTN. The T-coffee for ML171020.1 showed also better score and alingnment of sequences, and both Sec residues aligned together. Moreover, the phylogeny study indicated that the sequence found in that scaffold was significantly more closely related to the zebrafish SELENOP. The predicted protein was slightly smaller than the one found in zebrafish but shows great similarity.
Seblastian predicted a known selenoprotein (SelP) and a grade A SECIS element at the 3’UTR region.
Taking all these data into account, we were able to determine that in Anarrhichtys ocellatus, SELENOP is a selenoprotein. Its gene is located in the forward strand and it has 4 exons. The SECIS element is found downstream in the same strand.
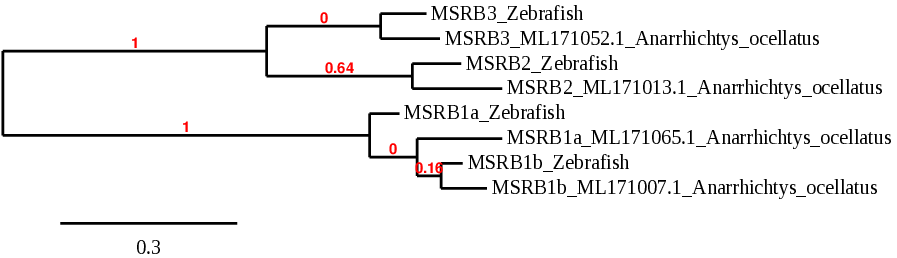
MSRB stand for Methionine Sulfoxide reductases. These proteins are thiol-dependent enzymes that catalyze the conversion of methionine sulfoxide to methionine. Their function is to repair enzymes that protect proteins from oxidative stress. This family includes 4 proteins: MSRB1a, MSRB1b, MSRB2 and MSRB3[1,25,26].
MSRB1 localizes to the cell nucleus and cytosol and it is expressed in a variety of adult and fetal tissues such as liver and kidney. In zebrafish, both MSRB1a and MSRB1b are selenoproteins.
Looking for MSRB1a in our studied species, we found two possible scaffolds. The scaffold ML171065.1 and the other candidate (ML171007.1) showed similar results in the TBLASTN and the Tcoffee, in which both scaffolds showed a good alignment and the presence of the Sec residue in the same position as zebrafish. However, when we analyzed the phylogenetics, we found that ML171065.1 was more closely related to the same protein in zebrafish, so we chose that scaffold. The predicted protein was 6 amino acids shorter than the one extracted from zebrafish, possibly due to a misprediction from exonerate. Seblastian predicted the selenoprotein MSRB1a and a grade A SECIS element in the 3’UTR region.
Considering these results, we were able to determine that Anarrhichtys ocellatus has a Sec-containing MSRB1a located in the reverse strand of the scaffold ML171065.1. Its encoding gene has 4 exons and a grade A SECIS element next to the first exon in the same strand, at the 3’UTR region.
Similarly to MSRB1a, MSRB1b showed the same two potential scaffolds. Eventhough the results from TBLASTN and T-coffee were very similar, phylogeny indicated that the closest related to MSRB1b in zebrafish was the scaffold ML171007.1. Tcoffee showed a good alignment and the presence of selenocysteine at the same location as the zebrafish.
We found that the structure of MSRB1b gene resembled MSRB1a, as it also had 4 exons, but in this case, in the forward strand. The predicted protein was almost identical to MSRB1a and Seblastian predicted the selenoprotein and a SECIS element in the 3’UTR region. The only thing that differed from MSRB1a was the location, as the two proteins are found in two different scaffolds and in different strands.
Previous studies suggest that several selenoprotein genes are duplicated in bony fishes such as zebrafish or Anarrhichtys ocellatus, probably due to the whole genome duplication that took place in fish early in evolution[5]. These studies indicate that, in some fish, MRB1b is a selenoprotein generated by that duplication event. To our knowledge, this duplication can be confirmed in Anarrhichtys ocellatus, as MSRB1a and MSRB1b show very similar sequence and structure but are found in different regions of the genome[5].
MSRB2 is expressed in a wide variety of tissues but its function is unknown. Studies show that overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage [26,9]. In zebrafish, MSRB2 is not a selenoprotein, it is a cysteine homologue.
For MSRB2, TBLASTN showed two possible scaffolds, but ML171013.1 had a lower E-value and greater identity than the other candidate. Furthermore, the phylogeny study revealed that this scaffold was more closely related to the same protein in zebrafish.
The Tcoffee showed a good alignment with a score of 999. The predicted protein was slightly smaller that the protein in zebrafish. We observed that the first 48 amino acids were missing in the predicted protein in Anarrhichtys ocellatus, including the methionine at the start. This finding could be due to a misprediction of the gene structure from exonerate. The predicted protein, like the zebrafish’s, did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
All these data demonstrate that in Anarrhichtys ocellatus, MSRB2 is a cys-containing homologue located in the scaffold ML171013.1 and its gene contains 4 exons in the reverse strand. There are no SECIS elements.
Unlike the other members of the family, MSRB3 acts as a monomer and requires zinc as a cofactor. It is also expressed in several different tissues and its function is not entirely known. In zebrafish, MSRB3 is not a selenoprotein, it is a cysteine homologue.
For MSRB3, TBLASTN showed four possible scaffolds. The scaffold ML171052.1 showed lower E-value and higher identity than the other candidates. Moreover, the phylogeny indicated that this scaffold had a closer origin to the same protein in zebrafish. The alignment using T-coffee had a high score and the predicted protein had almost the same size as the one found in zebrafish, only the last 6 nucleotides/amino acids were missing. Neither MSRB3 from zebrafish nor MSRB3 from Anarrhichtys ocellatus contain selenocysteine, consequently, Seblastian could not predict any selenoprotein or SECIS element.
All these data determine that in Anarrhichtys ocellatus, MSRB3 is a cys-containing homologue located in the scaffold ML171052.1 and its gene contains 6 exons in the forward strand. There are no SECIS elements.
This protein is a transmembrane protein located in the ER. Like SELENOK, Its functions involve: the ER-associated degradation of misfolded proteins, the reduction of disulfides in glycoprotein substrates and the mediation of anti-inflammatory effects of Se. In zebrafish, SELENOS is a selenoprotein[1].
TBLASTN for SELENOS showed a single scaffold candidate: ML171169.1. This scaffold had an acceptable E-value and identity. T-coffee showed a good alignment and the localization of Sec residue in both species (Anarrhichtys ocellatus and zebrafish) coincided . However, our predicted protein was missing the first 25 amino acids, including the first methionine. This could be due to a misprediction from exonerate. Seblastian was able to predict a known selenoprotein (SelS) and a grade A SECIS element in the 3’UTR region.
In summary, in Anarrhichtys ocellatus, SELENOS is a selenoprotein located in the scaffold ML171169.1. Its gene contains 5 exons in the reverse strand. The SECIS element is located in that same strand at the 3’UTR region.
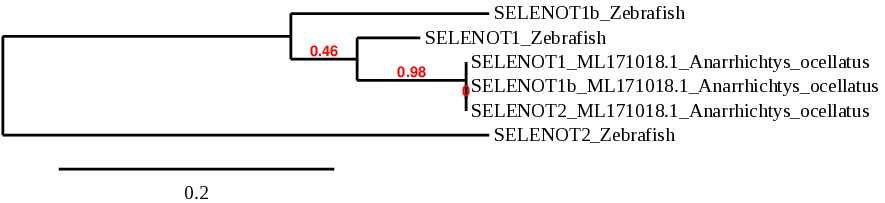
This protein is mainly localized to the ER and Golgi and it is ubiquitously expressed both during embryonic development and in adult tissues. The function of SELENOT is the regulation of calcium homeostasis and neuroendocrine function. In zebrafish, SELENOT proteins are selenoproteins[1].
Studies in fish selenoproteomes show that, as it has been mentioned before, some selenoproteins underwent duplication early in fish evolution. SELENOT is one of them. In zebrafish, SELENOT duplicated and generated SELENOT1b[5].
Interestingly, we found that in Anarrhichtys ocellatus’ genome, there is only one copy of SELENOT. When analyzing the proteins SELENOT1, SELENOT1b and SELENOT2 in our species of study, we observed that they were all the same protein.
The three proteins were found in the scaffold ML171018.1. That scaffold showed the lower E-value and higher identity in both SELENOT1 and SELENOT1b. That was not the case of SELENOT2, however, that scaffold obtained the best T-coffee alignment in the three SELENOT. Furthermore, the phylogeny assigned the three proteins the same exact position in the tree. When looking at the predicted protein sequence, we noticed that the sequences were identical, only differing in the first amino acids. The genes that encode the SELENOT proteins (which is actually a single gene) they all contained 5 exons in the same positions more or less (variating in approximately 20 amino acids at the beginning of exon 1) in the forward strand. Moreover, Seblastian predicted the same SECIS elements in the three fastasubseq files.
In order to discard a possible in duplication, we checked other scaffolds. Their T-coffes were way worse than ML171018.1. Moreover, the sequence around the X residue was essentially the same. It is possible that part of the protein has duplicated in some other scaffold, but based on our study we can not confirm that hypothesis.
Taking all that into consideration, we suggest that in Anarrhichtys ocellatus, the SELENOT family consists in only one protein found in scaffold ML171018.1. Its gene contains 5 exons in the forward strand and the SECIS element is in the 3’UTR region of the same strand.
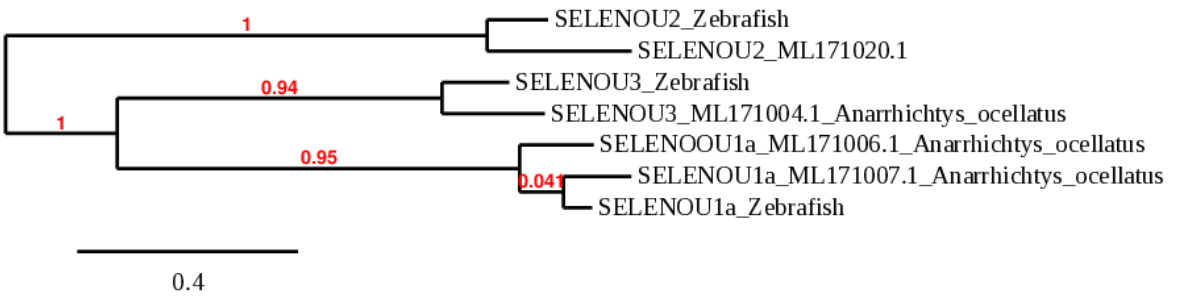
SELENOU proteins belong to the peroxiredoxin-like FAM312 superfamily. This family is widely distributed across the eukaryotic domain with either Cys- or Sec-containing proteins SELENOU expression in fish embryos is ubiquitous[1]. In zebrafish, SELENOU1a is a selenoprotein whereas SELENOU2 and SELENOU3 are cysteine homologues.
When analyzing SELENOU1a, TBLASTN showed two possible scaffolds. Even though the scaffold ML171006.1 had a lower E-value than ML171007.1, when running the T-coffee, ML171007.1 showed a significantly better alignment, as the coverage and the score were larger. Furthermore, the phylogeny indicated that ML171007.1 was considerably more related to SELENOU1a in zebrafish. The size of the predicted protein was almost the same as the homologue in zebrafish, only a few amino acids were missing at the beginning of the sequence. However, the predicted sequence started with methionine, the usual start amino acid for proteins.
Seblastian was able to predict a known selenoprotein (SelU1a) and multiple SECIS. Of those SECIS, only one was suitable for our protein when it came to location. There was a grade A SECIS at the 3’UTR region of our protein.
To sum up, in Anarrhichtys ocellatus, SELENOU1a is a selenoprotein located in the scaffold ML171007.1. Its gene contains 5 exons in the forward strand. The SECIS element is located in that same strand at the 3’UTR region.
For SELENOU2, TBLASTN showed a single possible scaffold, ML171020.1. The alignment using T-coffee had a high score and the predicted protein had the same size as the one found in zebrafish, with only a few missaligned amino acids. Neither SELENOU2 from zebrafish nor SELENOU2 from Anarrhichtys ocellatus contain selenocysteine, consequently, Seblastian could not predict any selenoprotein or SECIS element.
All these data determine that in Anarrhichtys ocellatus, SELENOU2 is a cys-containing homologue located in the scaffold ML171020.1 and its gene contains 6 exons in the reverse strand. There are no SECIS elements.
For SELENOU3, TBLASTN showed a single possible scaffold, ML171004.1. The alignment using T-coffee had a high score and the predicted protein had the same size as the one found in zebrafish, with only a few missaligned amino acids. Neither SELENOU3 from zebrafish nor SELENOU3 from Anarrhichtys ocellatus contain selenocysteine, consequently, Seblastian could not predict any selenoprotein or SECIS element.
All these data determine that in Anarrhichtys ocellatus, SELENOU3 is a cys-containing homologue located in the scaffold ML171004.1 and its gene contains 6 exons in the reverse strand. There are no SECIS elements.
SELENOW belongs to a group of proteins related to stress response. Its expression is regulated by the availability of Se in the diet and it seems that this protein could be involved in redox regulation of some proteins [28]. In zebrafish, SELENOW proteins are selenoproteins [9].
Similarly to SELENOO and SELENOT, SELENOW underwent duplication early in fish evolution. When analyzing the proteins SELENOW.1 and SELENOW.2 in our species of study, we observed that they were all the same protein.
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. Despite ML171011.1 was more closely related to the same protein in zebrafish when analysing phylogenetics, the scaffold ML171043.1 was chosen, because it showed better E-value and identity at TBLASTN than the other candidate. Moreover, the alignment of this scaffold using T-coffee showed better alignment and the predicted protein had the same size as the one found in zebrafish. Seblastian was not able to predict the selenoprotein SELENOW, but a grade A SECIS element in the 3’UTR region was found.
Considering these results, we were able to determine that Anarrhichtys ocellatus has a Sec-containing SELENOW located in the reverse strand of the scaffold ML171043.1. Its encoding gene has 4 exons and a SECIS element downstream in the same strand.
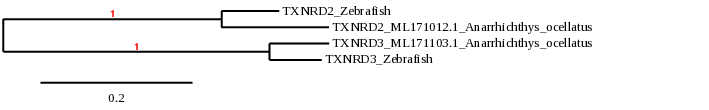
Thioredoxin reductase is the major intracellular protein disulfide reductant. It occurs in all organisms and is often an essential protein. In fish only two TXNRD forms are present, the mitochondrial TXNRD2 and an ancestral TXNRD3, from which the vertebrate cytosolic TRXND1 and testis-specific TXNRD3 evolved via a gene duplication event [29]. In zebrafish, TXNRD proteins are selenoproteins [9].
For TXNRD2, analysis in Anarrhichtys ocellatus showed four potential scaffolds. The scaffolds ML171103.1 and ML171012.1 showed lower E-value and higher identity than the other candidates. Eventhough the results from TBLASTN and T-coffee were very similar between both of them, the phylogeny indicated that the closest related to TXNRD2 in zebrafish was the scaffold ML171012.1. T-coffee showed a good alignment and the presence of selenocysteine at the same location as the zebrafish. Moreover, the predicted protein had the same size as the one found in zebrafish. Seblastian was not able to predict the selenoprotein TXNRD2, but a grade A SECIS element in the 3’UTR region was found.
All these data determine that in Anarrhichtys ocellatus, TXNRD2 is a selenoprotein located in the scaffold ML171012.1. Its gene contains 16 exons in the forward strand. The SECIS element is located in that same strand at the 3’UTR region.
For TXNRD3, TBLASTN showed six potential scaffolds. The scaffold ML171103.1 showed better E-value and identity than the other candidates. The T-coffee for this scaffold showed also better score and alingnment of sequences. Moreover, the phylogeny study indicated that the sequence found in that scaffold was significantly more closely related to the zebrafish TXNRD3, and the predicted protein had the same size as the one found in zebrafish. Seblastian was able to predict one known selenoprotein and a grade A SECIS element in the 3’UTR region.
Considering these results, we were able to determine that Anarrhichtys ocellatus has a Sec-containing TXNRD3 located in the reverse strand of the scaffold ML171103.1. Its encoding gene has 16 exons and a SECIS element downstream in the same strand.
eEFsec is a protein involved in the process of selenocysteine biosynthesis. This protein’s main function is to deliver charged tRNA(sec) to the A site of the ribosome, through binding to the Sec insertion sequence (SECIS) mRNA hairpin. Selenocysteyl-tRNA stabilizes the C-terminal domain of eEFsec[10]. The coupling effect is critical to preventing nonproductive decoding attempts and hence forms a basis for effective selenoprotein synthesis.
Zebrafish does not contain a selenocysteine in its eEFsec amino acid sequence. Thus, eEFsec is a cysteine homologue. In Anarrhichtys ocellatus, eEFSec is a cysteine homologue found in scaffold ML171004.1. We obtained a good E-value and identity at TBLASTN. The T-coffee showed also a good score for the alignment of sequences. The predicted protein had approximately the same length, as the Zebrafish protein sequence only had 5 amino acids more at the beggining. We found one SECIS element trough Seblastian, but we rejectected it as it was not located in the same strand, thus could not perform its function. Furthermore, we did not expect to find SECIS elements without the presence of a selenocysteine.
These data shows that in Anarrhichtys ocellatus, eEFsec is a cys-containing homologue located in the scaffold ML171004.1 and its gene contains 7 exons in the reverse strand. Accordingly, there are no SECIS elements.
PSTK is an enzyme involved in the translation system of Sec in eukaryotes and archaea. It discriminates Ser tRNA[Ser]Sec and marks it by transferring a phosphate group to the Ser, giving rise to PSer tRNA[Ser]Sec [15]. In zebrafish, PSTK is not a selenoprotein, it is a cysteine homologue [9].
When analyzing Anarrhichtys ocellatus, we found one possible scaffold (ML171007.1), which showed great e-value and identity at TBLASTN. The t-coffee also showed a 965 score for the alignment of sequences. The predicted protein missed 30 amino acids. This finding could be due to a misprediction of the gene structure from exonerate. The predicted protein, like the zebrafish homologue, did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
Taking this into account, in Anarrhichtys ocellatus, PSTK is a machinery cys-containing homologue protein. Its gene is located in the scaffold ML171007.1 and contains 3 exons in the reverse strand. It do not contain SECIS elements nor a Sec residue.
SBP2 is required for the co-translational incorporation of Sec at specific UGA codons. Their COOH-terminal RNA binding domain (RBD) and Sec incorporation domain (SID) form a complex that binds to the SECIS element. The RBD interacts with the SECIS, while SID enhances that interaction[16]. In zebrafish, SBP2 is not a selenoprotein, it is a cysteine homologue [9].
When analyzing Anarrhichtys ocellatus, we found two possible scaffolds. The scaffold ML171012.1 showed lower e-value and greater identity at TBLASTN than the other candidate (ML171011.1). The T-coffee also showed a good score for the alignment of sequences. The predicted protein had almost exactly the same length; the last 3 amino acids were missing. This finding could be due to a misprediction of the gene structure from exonerate. The predicted protein, like the zebrafish’s, did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
To summarize, SBP2 is a cys-containing homologue involved in Sec synthesis. It is located in the scaffold ML171012.1 and its gene contains 11 exons in the forward strand.
Selenocysteine synthase uses PSer tRNA[Ser]Sec and the active donor of selenium, selenophosphate, to form Sect RNA[Ser]Sec [15]. In zebrafish, SecS is not a selenoprotein, it is a cysteine homologue [9].
When analyzing Anarrhichtys ocellatus, we found one possible scaffold. The scaffold ML171014.1 showed great e-value and identity at TBLASTN. The t-coffee also showed a good score for the alignment of sequences. The predicted protein missed the last 26 amino acids. This finding could be due to a misprediction of the gene structure from exonerate. The predicted protein, like the zebrafish’s, did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
All these data demonstrate that in Anarrhichtys ocellatus, SecS is a cys-containing homologue enzyme related to Sec synthesis, located in the scaffold ML171014.1. Its gene contains 11 exons in the reverse strand.
SEPHS catalyzes the synthesis of the active selenium donor selenophosphate that is necessary for Sec biosynthesis. It was also proposed to have a role in autoregulation of selenoprotein synthesis[1].
Various eukaryotes contain two SEPHS paralogues, SEPHS1 and SEPHS2. Between the two isoforms in eukaryotes, only SEPHS2 shows catalytic activity during selenophosphate synthesis, while SEPHS1 play an essential role in regulating cellular physiology. However, SEPHS1 contains other amino acids such as Thr, Arg, Gly, or Leu at the catalytic domain, and SEPHS2 contains only a Sec [17]. In zebrafish, SEPHS2 is a selenoprotein, but SEPHS is a cysteine homologue[1].
When analyzing SEPHS in Anarrhichtys ocellatus, we found three possible scaffolds. The scaffold ML171005.1 showed a better e-value and identity at TBLASTN. The t-coffee also showed a better score for the alignment of sequences. Moreover, the phylogeny study indicated that the sequence found in that scaffold was significantly closer related to the zebrafish SEPHS [Fig. phylogeny]. The predicted protein had exactly the same length and, like the zebrafish homologue, it did not contain Sec residues. Because of this, Seblastian using the fastasubseq sequence could not predict any selenoprotein nor SECIS elements.
Taking this data into account, in Anarrhichtys ocellatus, SEPHS is a machinery protein, its gene is located in the reverse strand and it has 9 exons. It do not contain SECIS elements nor a Sec residue.
Related to SEPHS2, what we obtained is that the predicted sequences referred to the same scaffold and localization as the one for SEPHS. Apart from this, we were not able to find the Sec residue that could align with the one in zebrafish and, according to that, Seblastian could not predict any selenoprotein nor SECIS elements. As a result, we suggest that in Anarrhichtys ocellatus we can only observe one SEPHS protein.
SECp43 has a direct role in selenoprotein synthesis regulation through its involvement in methylation of tRNA[Ser]Sec [30]. In zebrafish, Secp43 is not a selenoprotein, it is a cysteine homologue [9].
When analyzing Anarrhichtys ocellatus, TBLASTN showed three possible scaffolds. The scaffolds ML171132.1 and ML171060.1 had both lower E-value. Although ML171060.1 had better score than the other one, ML171132.1 showed a significantly better coverage when analysing the alignment in T-coffee. Furthermore, the phylogeny study revealed that this scaffold was more closely related to the same protein in zebrafish. The predicted protein was slightly smaller that the protein in zebrafish. We observed that 47 amino acids were missing in the predicted protein in Anarrhichtys ocellatus. This finding could be due to a misprediction of the gene structure from exonerate. The predicted protein, like the zebrafish’s, did not contain selenocysteine, so Seblastian was not able to predict any selenoprotein or valid SECIS element.
All these data demonstrate that in Anarrhichtys ocellatus, SECp43 is a cys-containing homologue enzyme involved in Sec synthesis, located in the scaffold ML171032.1. Its gene contains 8 exons in the reverse strand.