Finding an explanation to our results
SELENOPROTEINS & CYSTEINE HOMOLOGUES
DIO
In this family there is a duplication of DIO3, which is common in all Teleosts, caused by the teleost-specific whole-genome duplication.
In all the four proteins of this family, four different scaffolds showed hits that surpassed the threshold of e-value greater than 0.01 that we established. This could be due to the fact that DIO proteins are very similar, so the protein used as a reference can be aligned with other regions that also encode for other Iodothyronine deiodinases.
In order to clarify which hit belonged to each reference sequence, a phylogenetic tree was built with the predicted sequences for all four scaffolds of all four DIO proteins. A prediction was removed before the creation of the tree because the sequence was too short and gave worse T-coffee alignments than the others. The eliminated predicted sequence was the one obtained at scaffold SSNR01000011.1 with the reference protein DIO3a from Zebrafish. The obtained tree can be seen below.
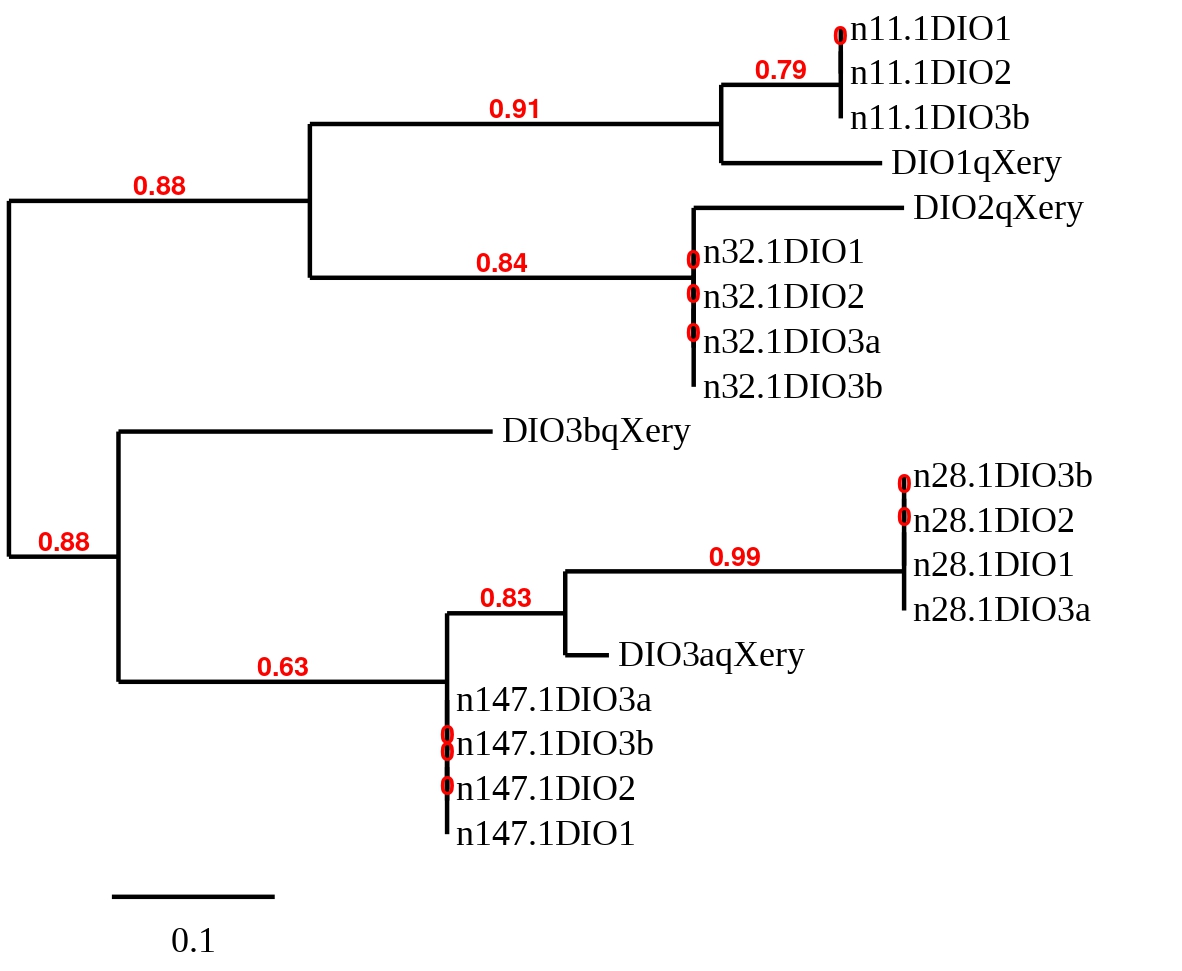
Taking into account the results obtained from the phylogenetic study the hit in scaffold SSNR01000011.1 was chosen for DIO1 from Zebrafish, the hit in SSNR01000032.1 for DIO2, the hit in SSNR01000032.1 for DIO3a and, finally, the one in SSNR01000147.1 for DIO3b. Then in order to clarify if the predicted sequences were selenoproteins, a detailed study was carried out.
DIO1
Six hits were found in this protein that surpassed the limit established (previously mentioned) when aligned with DIO1 from Danio rerio (SPP00000611_2.0). 3 belonged to scaffold SSNR01000011.1 (11.1), 1 to scaffold SSNR01000028.1 (28.1), 1 to SSNR01000032.1 (32.1) and 1 to SSNR01000147.1 (147.1).
As previously mentioned, scaffold 11.1 was chosen as closest to the query and was the only one in which Seblastian was performed.
The prediction found in this scaffold was located between positions 2707466 and 2709324 in the positive strand. It had 3 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a selenocysteine residue could be found in both compared sequences. On the other hand, the predicted protein didn’t have a Methionine (Met) at the beginning of the sequence.
When using Seblastian a better prediction was obtained because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction had 4 exons but the one obtained with the code only had 3. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. One SECIS element was predicted at the 3’-UTR end, situated between the positions 2710185-2710260 in the positive strand, 861 nucleotides after the end of the gene.
In conclusion, DIO1 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
DIO2
As happened with DIO1, 6 hits were found when aligning with the analogous protein from Danio rerio (SPP00000614_2.0). In this case, 2 belonged to scaffold SSNR01000011.1 (11.1), 1 to scaffold SSNR01000028.1 (28.1), 2 to SSNR01000032.1 (32.1) and 1 to SSNR01000147.1 (147.1).
The scaffold chosen as the best with the results collected from the phylogenetic tree was 32.1. The location of the gene was between 482998 and 480881 in the negative strand. It consisted of 3 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met amino acid couldn’t be. Nevertheless, the T-coffee alignment was considered as a good one, even though there were some gaps.
When testing with Seblastian a selenoprotein couldn’t be predicted. This could be due to the fact that the program uses a database where our protein is not included. Three SECIS elements were found but only two of them were located in the negative strand. One had a better grade than the other as indicated by Seblastian but was situated too far away from the 3’-UTR end of the predicted protein. And the other had a lower grade but was placed close enough to the 3’-UTR end. This last one could be found between positions 477038- 476967 and within a distance of 3914 nucleotides of 3’-UTR end. With this information, we considered this last potencial SECIS the correct one.
We reckon that DIO2 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
DIO3a
Differing from the two previous analysed proteins, 5 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000612_2.0). 2 belonged to scaffold SSNR01000011.1 (11.1), 1 to scaffold SSNR01000028.1 (28.1), 1 to SSNR01000032.1 (32.1) and 1 to SSNR01000147.1 (147.1).
As previously mentioned, scaffold 28.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in this scaffold was located between positions 4248679 and 4249422 in the positive strand. It only had 1 exon, that was situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
A better prediction was obtained with Seblastian because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction also had 1 exon and the length was similar between both predictions when comparing. One SECIS element was predicted at the 3’-UTR end, situated between positions 4249679-4249760 in the positive strand, 264 nucleotides after the end of the gene.
DIO3a from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
DIO3b
5 hits were found when aligning with the analogous protein from Danio rerio (SPP00000613_2.0). In this case, 2 belonged to scaffold SSNR01000011.1 (11.1), 1 to scaffold SSNR01000028.1 (28.1), 2 to SSNR01000032.1 (32.1) and 1 to SSNR01000147.1 (147.1).
The scaffold chosen as the best from the results collected from the phylogenetic tree was 147.1. The location of the gene was between 1099416 and 1098676 in the negative strand. It consisted in 2 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met couldn’t be in neither of the two compared proteins at the beginning of the sequence. Nevertheless, the T-coffee alignment could be considered as a good one.
When using Seblastian a better prediction was obtained because it had a few amino acids more at the N-terminal, including a Methionine as the first one. Two SECIS elements were found but only one of them was chosen by the program. It had a better grade and was situated between positions 1098347-1098275 and within a distance of 329 nucleotides of 3’-UTR end. With this information, we considered this potencial SECIS the correct one.
We conclude that DIO3b from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
Finally, to check whether the DIO proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, DIO1 from Zebrafish is very similar to the C.toddi’s chosen scaffold. The same applies to DIO2. Regarding DIO3a and DIO3b, the predicted DIO3b is more closely related to both DIO3a from Zebrafish and C.toddi.
GPx
The GPx family is composed by sequences that contain a selenocysteine residue while others are cysteine homologues.
Each protein from the GPx family when aligned to our species’ genome, showed various scaffolds with hits that passed the threshold established (previously mentioned). This could be due to the fact that GPx proteins are very similar, so the protein used as a reference can be aligned with other regions that also encode for other Glutathione Peroxidases.
In order to clarify which hit belonged to each reference sequence, a phylogenetic tree was built with the predicted sequences for all scaffolds of all GPx. Various predictions were removed before the creation of the tree because the sequences were too short and gave worse T-coffee alignments than the others or that a protein sequence couldn't be predicted. The eliminated predicted sequences were the ones obtained in scaffolds SSNR01000029.1 and SSNR01000140.1 when aligned with the reference protein GPx1a from Zebrafish; the ones from scaffolds SSNR01000029.1, SSNR01000127.1 and SSNR01000140.1 when aligned with GPx1b; the ones from scaffolds SSNR01000022.1 and SSNR01000140.1 when aligned with GPx2; the one from scaffold SSNR01000029.1 when aligned with GPx3a; the one from scaffold SSNR01000029.1 when aligned with GPx3b; the ones from scaffolds SSNR01000011.1, SSNR01000129.1 and SSNR01000222.1 when aligned with GPx4a, and finally the ones from scaffolds SSNR01000022.1, SSNR01000129.1 and SSNR01000222.1 when aligned with GPx4b. The obtained tree can be seen below.

Taking into account the results obtained from the phylogenetic study the hit in SSNR01000222.1 was chosen for GPx2, the hit in SSNR01000022.1 for GPx3a, the hit in SSNR01000183.1 for GPx3b, the hits in SSNR01000029.1 and SSNR01000140.1 for GPx4a and, finally, the one in SSNR01000127.1 for GPx4b. On the other hand, another phylogenetic tree was created in order to clarify which hit belonged to GPx1a and which to GPx1b (see below).

The results showed that the hit in scaffold SSNR01000085.1 belonged to GPx1a and the hit in scaffold SSNR01000129.1 belonged to GPx1b. Then to clarify if the predicted sequences were selenoproteins a detailed study was carried out.
GPx1a
14 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000621_2.0). 1 belonged to scaffold SSNR01000222.1 (222.1), 2 to scaffold SSNR01000085.1 (85.1), 3 to SSNR01000022.1 (22.1), 2 to SSNR01000129.1 (129.1), 2 to SSNR01000183.1 (183.1), 1 to SSNR01000029.1 (29.1), 1 to SSNR01000050.1 (50.1), 2 to SSNR01000127.1 (127.1) and 1 to SSNR01000140.1 (140.1).
Scaffolds 85.1 was chosen as closest to the query as mentioned above and was the only one where Seblastian was performed. It was located between positions 823692 and 822795 in the negative strand. It had 2 exons, that were situated as the following image shows.

The T-coffee alignment was also valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
The prediction obtained with Seblastian was as good as the one predicted by our program because it also didn’t include a Methionine as the first amino acid. This prediction also had 2 exons. One SECIS element was predicted at the 3’-UTR end, which was situated between the positions 822697-822626 in the negative strand, 98 nucleotides after the end of the gene. With this information, we can consider this one a possible SECIS candidate.
We suspect that GPx1a from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. We also believe that a duplication took place because the selenoprotein can be found in both scaffolds.
GPx1b
16 hits were detected when aligning the analogous protein from Danio rerio (SPP00000624_2.0). 2 belonged to scaffold SSNR01000222.1 (222.1), 2 to scaffold SSNR01000085.1 (85.1), 4 to SSNR01000022.1 (22.1), 2 to SSNR01000129.1 (129.1), 2 to SSNR01000183.1 (183.1), 1 to SSNR01000029.1 (29.1), 1 to SSNR01000050.1 (50.1), 2 to SSNR01000127.1 (127.1) and 1 to SSNR01000140.1 (140.1).
Scaffolds 129.1 was chosen as closest to the query and Seblastian was performed only on this one.
The prediction found in this scaffold was located between positions 294602 and 293845 in the negative strand. It had 2 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length and the predicted protein had a Met at the beginning of the sequence. Also, a selenocysteine residue could be found in both compared sequences.
The prediction obtained with Seblastian was as good as the one predicted by our program because it also included a Methionine as the first amino acid. This prediction also had 2 exons. One SECIS element was predicted at the 3’-UTR end, which was situated between the positions 293675-293605 in the negative strand, 170 nucleotides after the end of the gene. With this information, we can consider this one a potential SECIS candidate.
Taking everything into account, we suspect that GPx1b from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a possible SECIS element. We also believe that a duplication took place because the selenoprotein can be found in both scaffolds.
GPx2
12 hits were found when aligning with the analogous protein from Danio rerio (SPP00000616_2.0). In this case, 1 belonged to scaffold SSNR01000222.1 (222.1), 2 to scaffold SSNR01000129.1 (129.1), 2 to SSNR01000085.1 (85.1), 2 to SSNR01000083.1 (83.1), 1 to SSNR01000029.1 (29.1), 1 to SSNR01000127.1 (127.1), 2 to SSNR01000022.1 (22.1) and 1 to SSNR01000140.1 (140.1).
The scaffold chosen as the best from the results collected from the phylogenetic tree was 222.1. The location of the gene was between 797683 and 796952 in the negative strand. It consisted of 2 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected and a Met amino acid could be too. For this reason, the T-coffee alignment was considered appropriate.
A poorer prediction was obtained with Seblastian because it didn’t have a Methionine as the first one amino acid. This prediction also had 2 exons. One SECIS element was predicted at the 3’-UTR end, situated between positions 796874-796800 in the negative strand, 78 nucleotides after the end of the gene.
We reckon that GPx2 from Callopanchax toddi can be considered as a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
GPx3a
Differing from the previous analysed protein 14 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000617_2.0). 4 belonged to scaffold SSNR01000022.1 (22.1), 2 to scaffold SSNR01000085.1 (85.1), 2 to SSNR01000129.1 (129.1), 2 to SSNR01000183.1 (183.1), 2 to SSNR01000222.1 (222.1), 1 to SSNR01000029.1 (29.1) and 1 to SSNR01000050.1 (50.1).
As previously mentioned, scaffold 22.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in this scaffold was located between positions 2972334 and 2989352 in the positive strand. It had 5 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length, but the predicted protein didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
A better prediction was obtained with Seblastian because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction also had 5 exons and the length was similar between both predictions when comparing. One SECIS element was predicted at the 3’-UTR end, situated between the positions 2989452-2989529 in the positive strand, 100 nucleotides after the end of the gene.
GPx3a from Callopanchax toddi can be classified as a selenoprotein because it has a selenocysteine residue and a SECIS element. Also, it is very similar to the Zebrafish homologue, probably being orthologous genes.
GPx3b
13 hits were found when aligning with the analogous protein from Danio rerio (SPP00000623_2.0). In this case, 2 belonged to scaffold SSNR01000183.1 (183.1) , 2 to scaffold SSNR01000129.1 (129.1), 2 to SSNR01000085.1 (85.1), 2 to SSNR01000222.1 (222.1), 1 to SSNR01000029.1 (29.1), 3 to SSNR01000022.1 (22.1) and 1 to SSNR0100050.1 (50.1).
The scaffold chosen as the best from the results collected from the phylogenetic tree was 183.1. The location of the gene was between 702039 and 702817 in the positive strand. It consisted in 4 exons, which positions are indicated in the image below.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length and the predicted protein had a Met at the beginning of the sequence. Also, a selenocysteine residue could be found in both compared sequences.
A poorer prediction was obtained with Seblastian because it didn’t have a Methionine as the first one amino acid. This prediction also had 4 exons. One SECIS element was predicted at the 3’-UTR end, situated between the positions 702966-703047 in the negative strand, 149 nucleotides after the end of the gene.
We conclude that GPx3b from Callopanchax toddi is a selenoprotein. It is very similar to the Zebrafish homologue too.
GPx4a
10 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000618_2.0). 2 belonged to scaffold SSNR01000127.1 (127.1), 2 to scaffold SSNR01000140.1 (140.1), 1 to SSNR01000029.1 (29.1), 1 to SSNR01000050.1 (50.1), 1 to SSNR01000129.1 (129.1), 1 to SSNR01000085.1 (85.1), 1 to SSNR01000222.1 (222.1) and 1 to SSNR01000022.1 (22.1).
Scaffolds 29.1 and 140.1 were chosen as closest to the query as mentioned above and Seblastian was performed on both of them.
scaffold 29.1
Regarding the prediction found in the scaffold 29.1, it was located between positions 2540323 and 2540054 in the negative strand. It only had one exon, that was situated as the following image shows.

The T-coffee alignment was acceptable because the homology was relatively sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence. Besides, a selenocysteine residue found in the query sequence was a cysteine in the predicted sequence.
When testing with Seblastian a selenoprotein couldn’t be predicted. This could be, as we said before, due to the fact that the program uses a database where our protein is not included. Three SECIS elements were found but only two of them were located in the negative strand. Both of them had the same grade indicated by Seblastian but the locations in the genome were different. One was situated 11774 amino acids before the 3'-UTR end of the gene and the other was 45265 nucleotides before the same point. For this reason, both were situated too far away from the 3’-UTR end of the predicted protein. With this information, we considered that none of them could be a potential SECIS.
scaffold 140.1
The prediction found in this scaffold was located between positions 1007627 and 1010555 in the positive strand. It had 3 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Neither of the compared proteins had a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
When using Seblastian a similar prediction was obtained because it also lacked a Met as the first one amino acid. This prediction had 6 exons, which differs from the results obtained in our program. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. Two SECIS elements were predicted at the 3’-UTR end, one had a better grade than the other and was located between positions 1012630-1012714 in the positive strand, 2075 nucleotides after the end of the gene. So, with this information we can consider this one a possible SECIS candidate.
We suspect that GPx4a from Callopanchax toddi may be a selenoprotein because in one scaffold it has a selenocysteine residue but in the other this residue was lost and substituted by a cysteine. To confirm this hypothesis a final phylogenetic tree was performed (see at the end of the family)
GPx4b
Differing from the previous analysed protein 16 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000622_2.0). 4 belonged to scaffold SSNR01000127.1 (127.1), 3 to scaffold SSNR01000140.1 (140.1), 1 to SSNR01000029.1 (29.1), 1 to SSNR01000050.1 (50.1), 2 to SSNR01000129.1 (129.1), 2 to SSNR01000085.1 (85.1), 1 to SSNR01000022.1 (22.1) and 2 to SSNR01000222.1 (222.1).
As previously mentioned, scaffold 127.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in the scaffold 127.1 was located between positions 470400 and 469597 in the negative strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length, but the predicted protein didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
When using Seblastian a better prediction was obtained because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction had 7 exons, which differs from the results obtained in our program. One SECIS element was predicted at the 3’-UTR end, which was situated between the positions 470400-469597 in the negative strand, 479 nucleotides after the end of the gene. As what happened before with scaffold 140.1 from GPx4a, we can consider this one a possible SECIS candidate.
GPx4b from Callopanchax toddi can be classified as a selenoprotein because it has a selenocysteine residue and there is a possible SECIS element.
GPx7
7 hits were found in this protein when they were aligned with GPx7 from Danio rerio (SPP00000619_2.0). 2 belonged to scaffold SSNR01000029.1 (29.1), 2 to scaffold SSNR01000050.1 (50.1), 1 to SSNR01000022.1 (22.1), 1 to SSNR01000222.1 (222.1) and 1 to SSNR01000127.1 (127.1).
As previously mentioned, scaffold 29.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in the scaffold 29.1 was located between positions 2540314 and 2538599 in the negative strand. It had 2 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a cysteine residue could be found in both compared sequences, showing no differences between compared sequences. However, none of the proteins had a Methionine at the beginning of the sequence.
When testing with Seblastian a selenoprotein couldn’t be predicted. Three SECIS elements were found but only two of them were located in the negative strand. Both of them had the same grade indicated by Seblastian but both were situated in different positions. One could be found between positions 2528280-2528202 and within a distance of 10319 amino acid of 3’-UTR end and the other was situated between positions 2494789-2494705 and within a distance of 43810 nucleotides from 3’-UTR. With this information, we considered that none of them could be a potential SECIS element.
In conclusion, GPx7 from Callopanchax toddi can’t be a selenoprotein because it has a cysteine residue and there aren’t any SECIS element close enough. Also, it is very similar to the Zebrafish homolog, which it’s also not a selenoprotein. We could argue that they are orthologous genes.
GPx8
11 hits were found when aligning with the analogous protein from Danio rerio (SPP00000620_2.0). In this case, 3 to SSNR01000050.1 (50.1), 2 to SSNR01000029.1 (29.1), 1 to SSNR01000085.1 (85.1), 1 to scaffold SSNR01000129.1 (129.1), 1 belonged to scaffold SSNR01000222.1 (222.1), 1 to SSNR01000140.1 (140.1) and 1 to SSNR01000183.1 (183.1).
The scaffold chosen as the best from the results collected from the phylogenetic tree was 50.1. The location of the gene was between 2418827 and 2419768 in the positive strand. It consisted of 3 exons, which positions are indicated in the image below.

A cysteine residue could be found in both compared sequences, showing no differences between them but none of the proteins had a Methionine at the beginning of the sequence. However, the homology was sufficient for the proteins’ length to determine that the T-coffee alignment was considered appropriate.
Not one prediction of the protein and SECIS element could be obtained with Seblastian, which indicated that there were no SECIS element in this protein.
With all this information we reckon that GPx8 from Callopanchax toddi is not a selenoprotein because it doesn’t have any selenocysteine residue and SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
Finally, to check whether the GPx proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, all proteins from Zebrafish are very similar to the C.toddi’s chosen scaffold except the hit in scaffold 29.1 from GPx4a. We noticed that this hit corresponds to Zebrafish’s GPx7, this could be due to the fact that GPx proteins are very similar, so the protein used as a reference can be aligned with other regions that also encode for other GPx, in this case our predicted protein was aligned incorrectly to GPx4a from Zebrafish because of its similarity with GPx7, even though the last one is a cysteine homologue.
MSRB
This family is consisted by homologues MSRB1a, MSRB1b, MSRB2 and MSRB3 that have been annotated in Zebrafish
In order to clarify which hit belonged to each reference sequence, phylogenetic trees were built with the predicted sequences for all scaffolds of all MSRB proteins. The first tree corresponds to the possible selenoproteins and the second to the cysteines homologous. Two predictions were removed before the creation of the tree due to the fact that a sequence couldn’t be predicted. The eliminated predicted sequences were the ones obtained in scaffold SSNR01000008.1 with the reference proteins MSRB2 and MSRB3 from Zebrafish. The obtained trees can be seen below.


As previously mentioned, scaffold 104.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in the scaffold 104.1 was located between positions 1225316 and 1222966 in the negative strand. It had 5 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a selenocysteine residue and a Met, at the beginning of the sequence, could be found in both compared sequences.
When using Seblastian two predictions were obtained and both had 4 exons differing from the one predicted with the code. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. Four SECIS elements were predicted but Seblastian only considered two as a possible candidates. Each of these ones matched with the 3’-UTR end of each sequence, the first one was situated between positions 1224162-1224083 in the negative strand, in the middle of the gene. The other was situated between the positions 1222666-1222591, 300 nucleotides after the 3’-UTR of the gene. So, according to the literature used the second SECIS candidate was the correct one.
In conclusion, MSRB1a from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
MSRB1b
8 hits were found when aligning with the analogous protein from Danio rerio (SPP00000644_2.0). In this case, 1 belonged to scaffold SSNR01000008.1 (8.1) and 7 to scaffold SSNR01000104.1 (104.1).
The scaffold chosen as the best from the results collected from the phylogenetic tree was 8.1. The location of the gene was between 3283049 and 3281276 in the negative strand. It consisted of 3 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected as well as a Met amino acid. Also, the T-coffee alignment was considered as a good one.
When testing with Seblastian a selenoprotein could be predicted and the length of both predicted proteins was very similar. Two SECIS elements were found but only one of them was considered as a good enough candidate by Seblastian. It was found between positions 3280949-3280876 at the negative strand, within a distance of 327 nucleotides of 3’-UTR end. With this information, we considered this potencial SECIS the correct one.
We reckon that MSRB1b from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
MSRB2
In this case 7 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000646_2.0). 1 belonged to scaffold SSNR01000008.1 (8.1), 4 to scaffold SSNR01000068.1 (68.1), 1 to SSNR01000109.1 (109.1) and 1 to SSNR01002305.1 (2305.1).
As previously mentioned, scaffold 68.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in this scaffold was located between positions 1598962 and 1599964 in the positive strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence and a few amino acids were missing as well. And, due to the fact that MSRB2 is a cysteine homologue, a selenocysteine residue couldn’t be found in neither compared sequences.
No SECIS structure or protein prediction were obtained when using Seblastian.
All the information gathered confirms that MSRB2 from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue nor a SECIS element but it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
MSRB3
13 hits were found when aligning the analogous protein from Danio rerio (SPP00000647_2.0). In this case, 1 belonged to scaffold SSNR01000008.1 (8.1) , 3 to scaffold SSNR01000068.1 (68.1), 5 to SSNR01000109.1 (109.1) and 4 to SSNR01002305.1 (2305.1).
The scaffolds chosen as the bests of the results collected from the phylogenetic tree were 109.1 and 2305.1.
scaffold 109.1
The location of the gene was between 1311474 and 1315752 in the positive strand. It consisted in 6 exons, which positions are indicated in the image below.

Due to the fact that MSRB3 is a cysteine homologue a selenocysteine residue couldn’t be detected but a Met could in both of the two compared proteins at the beginning of the sequence. All in all, the T-coffee alignment could be considered as a good one.
When using Seblastian two SECIS were predicted. Both SECIS were located in the negative strand so neither of them were considered as possible SECIS elements for this gene.
scaffold 2305.1
The location of the gene was between 1311474 and 1315752 in the positive strand. It consisted in 6 exons, which positions are indicated in the image below.

Due to the fact that MSRB3 is a cysteine homologue a selenocysteine residue couldn’t be detected and neither could a Met amino acid at the beginning of the sequence. All in all, the T-coffee alignment could be considered as a good one, even though a few amino acids of the beginning of the predicted sequence are missing.
No SECIS structure or protein prediction were obtained when using Seblastian.
We conclude that MSRB3 from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue and neither has a SECIS element. In this case we can see that a duplication has occured because we find the whole protein in two different scaffolds.
Finally, to check whether the MSRB proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, each protein from Zebrafish is organised together with the homologue from Callopanchax toddi, corroborating that the assignment for each genomic region was correct. Also, we can see that MSRB2 and MSRB3 are more similar between them than with MSRB1a and MSRB1b, this is consistent with the fact that the two first proteins are cysteine homologues while the last two are selenoproteins. We also believe that a duplication of MSRB3 has taken place because it can be found in two different scaffolds.
Sel15
3 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000632_2.0). All 3 belonged to scaffold SSNR01000598.1 (598.1) so it was the one were Seblastian was performed.
Regarding the prediction found in this scaffold, it was located between positions 57695 and 52075 in the negative strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was also valid because the homology was sufficient for the proteins’ length. Neither of the compared sequences had a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both.
The prediction obtained with Seblastian was better than the one predicted by our program because it included a Methionine as the first amino acid. This prediction had 5 exons whereas the one obtained with the code only had 4. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. One SECIS element was predicted at the 3’-UTR end, situated between positions 51672-51597 in the negative strand, 403 nucleotides after the end of the gene.
We conclude that Sel15 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoE
In this protein 3 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000617_2.0). 2 belonged to scaffold SSNR01000071.1 (71.1) and the other 2 to scaffold SSNR01003313.1 (3313.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created.

As seen in the tree, the prediction from scaffold 3313.1 is more similar to SelenoE from Zebrafish than the one from scaffold 71.1. So, scaffold 3313.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in this scaffold was located between positions 1144 and 2320 in the positive strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length but the predicted protein had a few amino acids more before the Met that corresponded to the first amino acid of the query. However, a selenocysteine residue could be found in both compared sequences.
A similar prediction was obtained with Seblastian. This prediction also had 4 exons and the length was similar between both predictions when comparing. One SECIS element was predicted at the 3’-UTR end, situated between the positions 2349-2440 in the positive strand, 29 nucleotides after the end of the gene.
SelenoE from Callopanchax toddi may not be classified as a selenoprotein because it has a selenocysteine residue and a SECIS element but it is too close to the end of the gene. However it is very similar to the Zebrafish homolog.
SelenoH
Two hits were found in this protein that surpassed the limit established (previously mentioned) when they were aligned with SelenoH from Danio rerio (SPP00000634_2.0). The two of them belonged to the same scaffold, SSNR01000176.1 (176.1). Seblastian was performed on this one predicted sequence.
The prediction found in the scaffold 176.1 was located between positions 509233 and 506869 in the negative strand. It had 3 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a selenocysteine residue could be found in both compared sequences. On the other hand, the predicted protein didn’t have a Met at the beginning of the sequence.
When using Seblastian a similar prediction was obtained because it also didn’t have a Methionine as the first amino acid. This prediction also had 3 exons and a similar length. One SECIS element was predicted at the 3’-UTR end, situated between the positions 2710185-2710260 in the positive strand, 206 nucleotides after the end of the gene.
In conclusion, SelenoH from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoI
10 hits were found when aligning with the analogous protein from Danio rerio (SPP00000635_2.0). In this case, 2 belonged to scaffold SSNR01000021.1 (21.1), 5 to scaffold SSNR01000055.1 (55.1) and 1 to SSNR01000637.1 (637.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created.

As seen in the tree, the prediction from scaffold 55.1 is more similar to SelenoI from Zebrafish than the ones from scaffolds 21.1 and 637.1. So, scaffold 55.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The location of the gene was between 1488201 and 1495976 in the positive strand. It consisted of 10 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met amino acid couldn’t be. Nevertheless, the T-coffee alignment was considered as a good one.
When testing with Seblastian a selenoprotein was predicted. The sequence predicted by Seblastian was better than the one predicted by the code because it has a Met as the first amino acid. A SECIS element was found between positions 1497011- 1497094 and within a distance of 1035 nucleotides of 3’-UTR end.
We reckon that SelenoI from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoJ1
12 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000236_2.0). 6 belonged to scaffold SSNR01000141.1 (141.1) and the other 6 to scaffold SSNR01000266.1 (266.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created.

As seen in the tree, the prediction from scaffold 266.1 is more similar to SelenoJ1 from Zebrafish than the one from scaffold 141.1. So, scaffold 266.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in the chosen scaffold was located between positions 364028 and 374801 in the positive strand. It had 9 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a M at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
A better prediction was obtained with Seblastian because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction also had 9 exons and the length was similar between both predictions when comparing. Six SECIS elements were predicted at the 3’-UTR end, but Seblastian considered one as the best. It was situated between the positions 374838-374912 in the positive strand, 37 nucleotides after the end of the gene. The other possible SECIS were checked but four of them were located in the negative strand and the last one could be found in the middle of the gene.
SelenoJ1 from Callopanchax toddi is not selenoprotein because it has a selenocysteine residue and but it doesn’t have a SECIS element, even though it is very similar to the Zebrafish homolog.
SelenoK
Two hits were found in this protein that surpassed the limit established (previously mentioned) when aligned with MSRB1a from Danio rerio (SPP00000633_2.0). All of them belonged to scaffold SSNR010000052.1 (52.1). It was the only one where Seblastian was performed.
The prediction found in the scaffold 52.1 was located between positions 2925848 and 2928149 in the positive strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a selenocysteine residue and a Met at the beginning of the sequence could be found in both compared sequences.
When testing with Seblastian a selenoprotein couldn’t be predicted. This could be due to the fact that the program uses a database where our protein is not included. Three SECIS elements were found and all were located in the positive strand. The first one had a high grade and it started in position 2933286 and ended in position 2933269, 5037 nucleotides after the end of the gene. The second one also had a high grade and started in position 2929596 and ended in position 2929679, 1447 nucleotides after the end of the gene. And finally the third one had the lowest grade, starting in position 2969713 and ending in position 2969770, 41564 positions after the 3’-UTR end of the gene. This predicted proteins had 2 possible SECIS elements candidates.
In conclusion, SelenoK from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoL
3 hits were found when aligning with the analogous protein from Danio rerio (SPP00000637_2.0). In this case, all of them belonged to scaffold SSNR01000243.1.
The location of the gene was between 138515 and 141829 in the positive strand. It consisted of 4 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met amino acid couldn’t be, this could be due to the fact that the sequence predicted had a few amino acids missing at the beginning. However, the T-coffee alignment was considered as a good one.
When testing with Seblastian a selenoprotein could be predicted but it had 8 exons. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. Three SECIS elements were found but only one of them was considered as a good enough candidate by Seblastian. It was found between positions 141898-141975 at the positive strand, and within a distance of 69 nucleotides of 3’-UTR end. With this information, we considered this potencial SECIS the correct one.
We reckon that SelenoL from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoM
3 hits were found when aligning with the analogous protein from Danio rerio (SPP00000638_2.0). In this case, 2 belonged to scaffold SSNR01000071.1 (71.1) and 1 to SSNR01003313.1 (3313.1).
Scaffold 71.1 was chosen as the only valid one because a protein couldn’t be predicted with scaffold 3313.1. So, it was the only one were Seblastian was performed.
The location of the gene was between 1002912 and 1004895 in the positive strand. It consisted in 5 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met couldn’t be in neither of the two compared proteins at the beginning of the sequence. Nevertheless, the T-coffee alignment was considered as a good one.
When using Seblastian a better prediction was obtained because it had a few amino acids more at the N-terminal, including a Methionine as the first one. Four SECIS elements were found but only one of them was chosen by the program. It had a better grade and was situated between positions 1005240-1005323 and within a distance of 245 nucleotides of 3’-UTR end. With this information, we considered this potencial SECIS the correct one.
We conclude that SelenoM from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoN
11 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000639_2.0). All of them belonged to scaffold SSNR01000028.1 (28.1).
The prediction found in the scaffold 28.1 was located between positions 3990349 and 3986703 in the negative strand. It had 12 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Both compared sequences had a Met at the beginning of the sequence as well as a selenocysteine residue at the middle.
A similar prediction was obtained with Seblastian. This prediction also had 12 exons and the length was similar between both predictions when comparing. One SECIS element was predicted at the 3’-UTR, situated between the positions 3985236-3985087 in the positive strand, 1466 nucleotides after the end of the gene.
SelenoN from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoO
SelenoO1
14 hits were found in this protein that surpassed the limit established (previously mentioned) when aligned with SelenoO from Danio rerio (SPP00000640_2.0). All of them belonged to scaffold SSNR01000209.1 (209.1).
The prediction found in this scaffold was located between positions 826635 and 843245 in the positive strand. It had 18 exons, that were situated as the following image shows.

The T-coffee alignment was valid, even though quite a few amino acids were added at the beginning of the sequence. Two selenocysteine residues could be found in the predicted protein while only one could be found in the query. A Met amino acid (as the first one) was found in both compared sequences.
When using Seblastian two predictions were obtained each one composed by 9 exons. This demonstrates that a duplication may have occurred in our specie, and the number of exons have doubled. Two SECIS elements were predicted at the 3’-UTR end. The first one was situated between positions 843659-843744 in the positive strand, 414 nucleotides after the end of the gene. The other one was also located in the positive strand, between positions 830517-830602 in the middle of the gene. In the end, only the first SECIS element is the only possible candidate.
In conclusion, SelenoO1 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. We believe that a duplication of this protein may have taken place in our specie.
SelenoO2
As happened with SelenoO1, 14 hits were found when aligning with the analogous protein from Danio rerio (SPP00000641_2.0). All of them belonged to scaffold SSNR01000209.1 (209.1).
The prediction found in this scaffold was located between positions 826725 and 843245 in the positive strand. It had 18 exons, that were situated as the following image shows.

The T-coffee alignment was valid, even though quite a few amino acids were added at the end of the sequence. One selenocysteine residue could be found in the predicted protein and in the query in different positions. A Met amino acid (as the first one) wasn’t found in neither of the compared sequences.
When using Seblastian two predictions were obtained each one composed by 9 exons. This demonstrates that a duplication may have occurred in our specie, and the number of exons have doubled. Two SECIS elements were predicted at the 3’-UTR end. The first one was situated between positions 843659-843744 in the positive strand, 414 nucleotides after the end of the gene. The other one was also located in the positive strand, between positions 830517-830602 in the middle of the gene. In the end, only the first SECIS element is the only possible candidate.
In conclusion, SelenoO2 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. We believe that a duplication of this protein may have taken place in our specie.
Finally, to check whether the SelenoO proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, SelenoO2 from Zebrafish has disappeared in C.toddi, because both predictions are closer to SelenoO1. Also, a duplication of SelenoO1 has taken place in our specie because because the number of exons has doubled and two selenocysteine residues can be found.
SelenoP
Five hits were found in this protein that surpassed the limit established (previously mentioned) when they were aligned with DIO1 from Danio rerio (SPP00000643_2.0). 2 belonged to scaffold SSNR01000008.1 (8.1) and 3 to scaffold SSNR01000009.1 (9.1).
Scaffold 9.1 was chosen as the only valid one because a protein couldn’t be predicted with scaffold 8.1. So, it was the only one were Seblastian was performed.
The prediction found in the scaffold 9.1 was located between positions 24060 and 23195 in the negative strand. It had 3 exons, that were situated as the following image shows.

A selenocysteine residue could be found in both compared sequences as well as a Met at the beginning of the sequences. The T-coffee alignment was valid because the homology was sufficient for the proteins’ length, even though various amino acids were missing at the end of the predicted sequence.
When using Seblastian two predictions were obtained each of them comprised 5 exons. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. The second sequence predicted by Seblastian was the most similar to the one predicted with the code. A SECIS element was predicted for each sequence at the 3’-UTR end. The one for the first sequence was situated between positions 21576-21499 in the negative strand, 1619 nucleotides after the end of the gene. And the one predicted for the other sequence was situated between positions 22096-22021 in the negative strand as well, 1099 nucleotides after the end of the gene. Both SECIS elements are valid candidates.
In conclusion, SelenoP from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoS
Only one hit was found when aligning with the analogous protein from Danio rerio (SPP00000648_2.0). It belonged to scaffold SSNR01000911.1 (911.1), and was the one were Seblastian was performed.
The location of the gene was between 49755 and 45051 in the negative strand. It consisted of 5 exons, which positions are indicated in the image below.

Four selenocysteines could be detected but only one matched with Zebrafish’s protein. The predicted protein didn’t start with a methionine differing from the reference protein. Nevertheless, the T-coffee alignment was considered as a good one, even though there were some gaps at the beginning.
When testing with Seblastian a selenoprotein couldn’t be predicted. This could be due to the fact that the program uses a database where our protein is not included. A SECIS element was found between positions 44540- 44460 in the negative strand, and within a distance of 511 nucleotides of 3’-UTR end.
We reckon that SelenoS from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoT
SelenoT1
5 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000649_2.0). All of them belonged to scaffold SSNR01000216.1 (216.1) so it was the only one were Seblastian was performed..
The prediction found in the scaffold 216.1 was located between positions 1012399 and 1006952 in the negative strand. It had 5 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence because a few amino acids were missing. However, a selenocysteine residue could be found in both compared sequences.
A better prediction was obtained with Seblastian because it had a few amino acids more at the N-terminal, including a Methionine as the first one. This prediction also had 5 exons and the length was similar between both predictions when comparing. One SECIS element was predicted at the 3’-UTR end, situated between positions 1006739-1006665 in the negative strand, 213 nucleotides after the end of the gene.
SelenoT from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoT1b
As it happened with SelenoT, 5 hits were found when aligning the analogous protein from Danio rerio (SPP00000651_2.0). Once again, all belonged to scaffold SSNR01000216.1 (216.1).
The location of the gene was between 1012342 and 1006907 in the negative strand. It had 5 exons, which positions are indicated in the image below.

A selenocysteine residue could be detected but a Met couldn’t be in the predicted sequence because a few of the firsts amino acids were missing. Nevertheless, the T-coffee alignment could be considered as a good one.
When using Seblastian a better prediction was obtained because it had a few amino acids more at the N-terminal, including a Methionine as the first one. One SECIS element was found between positions 1006739-1006665, the same ones as the SECIS element predicted for SelenoT, and within a distance of 168 nucleotides of 3’-UTR.
We conclude that SelenoT1b from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoT2
In this case, four hits were found in this protein that surpassed the limit established (previously mentioned) when aligned with SelenoT2 from Danio rerio (SPP00000650_2.0). But once again all of them belonged to scaffold SSNR01000216.1 (216.1).
The prediction found in the scaffold 216.1 was located between positions 1012342 and 1006931 in the negative strand. It had 5 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Also, a selenocysteine residue could be found but a Met at the beginning of the sequence couldn’t be because a few amino acids from the beginning were missing.
When using Seblastian a better prediction was obtained because it had the amino acids from the beginning of the sequence that the one predicted with the code lacked. A SECIS element was predicted with the same localization in the negative strand as the other two, 1006739-1006665, 192 nucleotides after the end 3’-UTR.
In conclusion, SelenoT2 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
Finally, to check whether SelenoT proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the results obtained from the tree, we can see that all the predicted sequences correspond to SelenoT1 from Zebrafish. SelenoT1b and SelenoT2 have been lost in our specie.
SelenoU
SelenoU1a
9 hits were found when aligning with the analogous protein from Danio rerio (SPP00000652_2.0). In this case, 6 belonged to scaffold SSNR01000024.1 (24.1) and 3 to scaffold SSNR01000386.1 (386.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created

As seen in the tree, the prediction from scaffold 24.1 is more similar to SelenoU1a from Zebrafish than the one from scaffold 386.1. So, scaffold 24.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The location of the gene was between 1738832 and 1744612 in the positive strand. It consisted of 9 exons, which positions are indicated in the image below.

In the predicted protein a selenocysteine residue could be detected but a Met amino acid at the beginning couldn't be in neither of the compared sequences. The T-coffee alignment was considered as a good one, though some amino acids from the start were lost.
When testing with Seblastian a selenoprotein couldn’t be predicted. This could be due to the fact that the program uses a database where our protein is not included. Three SECIS elements were found all of them located in the positive strand. The first one had a high grade and was between positions 1745025-1745096, 413 nucleotides after 3’-UTR end. The other two weren’t good candidates because one was found in the middle of the sequence and the other before the start of the sequence. So, with this information, we considered the first potencial SECIS the correct one.
We reckon that SelenoU1a from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SelenoU2
2 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000655_2.0). Both of them belonged to scaffold SSNR01000050.1 (50.1).
The prediction found in this scaffold was located between positions 289957 and 291196 in the positive strand. It only had 6 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. None of the two compared sequences started with a methionine. And, due to the fact that SelenoU2 is a cysteine homologue, a selenocysteine residue couldn’t be found in neither compared sequences.
A SECIS structure was obtained but a protein prediction wasn’t when using Seblastian. The SECIS element was located in the negative strand, so it couldn’t be a valid candidate.
All the information gathered confirms that SelenoU2 from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue nor a SECIS element but it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SelenoU3
4 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000656_2.0). All of them belonged to scaffold SSNR01000086.1 (86.1).
The prediction found in the scaffold 28.1 was located between positions 1007014 and 1003006 in the negative strand. It had 6 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Neither of the two compared sequences started with a methionine. And, due to the fact that SelenoU3 is a cysteine homologue, a selenocysteine residue couldn’t be found in neither compared sequences.
A SECIS structure was obtained but a protein prediction wasn’t when using Seblastian. The SECIS element was located in the negative strand, but in the middle of the gene, so it couldn’t be a valid candidate.
All the information gathered confirms that SelenoU3 from Callopanchax toddi is a cystein homologue because it doesn’t have a selenocysteine residue nor a SECIS element but it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
Finally, to check whether SelenoU proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, each protein from Zebrafish is organised together with the homologue from Callopanchax toddi, corroborating that the assignment for each genomic region was correct. SelenoU3 is evolutionary closer to SelenoU1a than with Seleno U2, which is unexpected because SelenoU2 and SelenoU3 are both cysteine hologues.
SelenoW
In this case, 3 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000658_2.0). All of them belonged to scaffold SSNR01000030.1 (30.1).
The prediction found in this scaffold was located between positions 3898081 and 3903415 in the positive strand. It had 2 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length, although some amino acids were missing at the beginning and at the end of the predicted protein. It also didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
Two predictions were obtained with Seblastian, one only had one exon while the other had 2. This could be caused by the fact that the predicted protein by the code was compared with two different species that weren’t Zebrafish. One SECIS element was predicted at the 3’-UTR end for each of the two predicted proteins. The first one was situated between positions 3903721-3903796, 306 nucleotides after the end of the gene. The other one was situated in the middle of the gene so it wasn’t considered as a possible SECIS element.
SelenoW from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a SECIS element and it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
TXNRD
In order to clarify which hit belonged to each reference sequence, a phylogenetic tree was built with the predicted sequences for all scaffolds of all TXNRD proteins. Different predictions were removed before the creation of the tree due to the fact that the sequences were too short and gave worse T-coffee alignments than the others. The tree obtained can be seen below.

As seen in the tree, the prediction from scaffolds 64.1 and 187.1 are more similar to TXNRD2 and TXNRD3 from Zebrafish than the ones from the other scaffolds. After, the T-coffee alignments were checked and predictions in scaffold 187.1 were really bad for both proteins, so it was removed. Finally only scaffold 64.1 was chosen as the only possible candidate.
TXNRD2
23 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000660_2.0). 11 belonged to scaffold SSNR01000064.1 (64.1), 5 to scaffold SSNR01000565.1 (565.1), 1 to SSNR01001382.1 (1382.1), 2 to SSNR01000187.1 (187.1), 2 to SSNR01000108.1 (108.1) and 1 to SSNR01002615.1 (2615.1).
As mentioned above scaffold 64.1 was the only selected hit. It was located between positions 2028300 and 2048272 in the positive strand. It had 15 exons, that were situated as the following image shows.

The T-coffee alignment was also valid because the homology was sufficient for the proteins’ length. The predicted protein didn’t have a Met at the beginning of the sequence. However, a selenocysteine residue could be found in both compared sequences.
The prediction obtained with Seblastian was as good as the one predicted by our program because it also didn’t include a Methionine as the first amino acid. This prediction also had 15 exons. Four SECIS elements were predicted at the 3’-UTR end but only one was selected as a possible SECIS element from Seblastian. It was situated between the positions 2050066-2050139 in the positive strand, 1794 nucleotides after the end of the gene. With this information, we can classify this one as a possible SECIS candidate.
Taking everything into account, we suspect that TXNRD2 from Callopanchax toddi is a selenoprotein because in their possible scaffolds it has a selenocysteine residue and a possible SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
TXNRD3
21 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000661_2.0). 10 belonged to scaffold SSNR01000565.1 (565.1) , 1 to scaffold SSNR010001382.1 (1382.1), 5 to SSNR01000064.1 (64.1), 1 to SSNR01000187.1 (187.1), 1 to SSNR01002615.1 (2615.1), 1 to SSNR01000003.1 (3.1), 1 to SSNR01000108.1 (108.1) and 1 to SSNR01000074.1 (74.1)
Scaffolds 64.1 was chosen as closest to the query and was the only one where Seblastian was performed. It was located between positions 2028297 and 2048272 in the positive strand. It had 11 exons, that were situated as the following image shows.

The T-coffee alignment was valid even though some gaps were present. The predicted protein didn’t have a Met at the beginning of the sequence and some amino acids were missing at the at the beginning. However, a selenocysteine residue could be found in both compared sequences.
The prediction obtained with Seblastian was better because it had 15 exons. This could be due to the fact that Seblastian uses a different reference genome that isn’t Zebrafish, so the results may vary. Four SECIS elements were predicted at the 3’-UTR end but only one was selected as a possible SECIS element by Seblastian. It was situated between the positions 2050066-2050139 in the positive strand, 1794 nucleotides after the end of the gene. With this information, we classified this one as a possible SECIS candidate.
Taking everything into account, we suspect that TXNRD3 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a possible SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
Finally, to check whether TXNRD proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, we can observe that the scaffolds 64.1 from TXNRD2 and TXNRD3 are more similar to TXNRD2 from Zebrafish than to TXNRD3. So, we believe that selenoprotein TXNRD3 has been lost in our specie.
MsrA
9 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000025_2.0). 4 belonged to scaffold SSNR01000062.1 (62.1), 4 to SSNR01000277.1 (277.1) and the last one to SSNR01001156.1 (1156.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created

As seen in the tree, the prediction from scaffold 277.1 is more similar to MsrA from Zebrafish than the ones from scaffolds 62.1 and 1156.1. So, scaffold 277.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The prediction found in the scaffold 277.1 was located between positions 377662 and 404852 in the positive strand. It had 4 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. Neither of the two compared sequences started with a methionine. And, due to the fact that MsrA is a cysteine homologue, a selenocysteine residue couldn’t be found in neither compared sequences.
Four SECIS structures were obtained but a protein prediction wasn’t when using Seblastian. All of them were located in the positive strand, but two of them in the middle of the gene, and the other two before the start of the gene. So none of them could be a valid candidate.
All the information gathered confirms that MsrA from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue nor a SECIS element but it is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
SELENOPROTEIN MACHINERY
SEPHS or SPS
This family is comprised by both a selenocysteine and a cysteine homologue
SEPHS1 or SPS1
9 hits were found when aligning with the analogous protein from Danio rerio (SPP00000629_2.0). In this case, 7 belonged to scaffold SSNR01000035.1 (35.1) and 8 to scaffold SSNR01000045.1 (45.1).
In order to choose which hit was the best t-coffee alignments were compared and a phylogenetic tree was created

As seen in the tree, the prediction from scaffold 35.1 is more similar to SEPHS1 from Zebrafish than the one from scaffold 45.1. So, scaffold 35.1 was chosen as closest to the query and was the only one were Seblastian was performed.
The location of the gene was between 641436 and 645112 in the positive strand. It consisted of 9 exons, which positions are indicated in the image below.

Looking T-coffee, which was considered as a good one, a Met amino acid at the beginning of the compared sequences could be found but selenocysteine residue couldn’t be detected because the two sequences were cysteines homologs.
When testing with Seblastian a selenoprotein couldn’t be predicted. Only one SECIS element was found located in the positive strand. It was between positions 690008-690071, 44896 nucleotides after 3’-UTR end. This SECIS element was not a good candidate because it was located too far from the end of the gene.
We reckon that SEPHS1 from Callopanchax toddi is a cysteine homologous because it hasn’t got a selenocysteine residue nor a possible SECIS element. Due to the fact that it is very similar to the Zebrafish homolog, we could speculate that they are orthologous genes.
SEPHS2 or SPS2
14 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000630_2.0). In this case, 6 belonged to scaffold SSNR01000035.1 (35.1) and 8 to scaffold SSNR01000045.1 (45.1).

The prediction found in the scaffold 45.1 was chosen due to closeness detected in the phylogenetic tree. It was located between positions 3347832 and 3354156 in the positive strand and it had 8 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length. The two compared sequences started with a methionine and a selenocysteine residue could be found in both of them.
When testing with Seblastian a selenoprotein could be predicted. Two SECIS structures were obtained but only one of them was considered good enough by the program with the maximum grade. The SECIS element was located from 3354524 to 3354605 nucleotides in the positive strand, 368 nucleotides after 3'-UTR region of the gene.
All the information gathered confirms that SEPHS2 from Callopanchax toddi is a selenoprotein because it has a selenocysteine residue and a potential SECIS element and is very similar to the Zebrafish homolog. We could argue that they are orthologous genes.
Finally, to check whether SEPHS proteins predicted in Callopanchax toddi are similar to their homologues in Danio rerio a phylogenetic tree was created (see below):

Taking into account the phylogenetic tree, each protein from Zebrafish is organised together with the homologue from Callopanchax toddi, corroborating that the assignment for each genomic region was correct. SEPHS1 from Zebrafish is very similar to the C.toddi’s chosen scaffold. The same applies to SEPHS2. We have to emphasize that these two proteins could also be denominated SPS1 and SPS2 respectively.
PSTK
In this case, 4 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000052_2.0). All of them belonged to scaffold SSNR01000183.1 (183.1).
The prediction found in this scaffold was located between positions 419826 and 418755 in the negative strand. It had 3 exons, that were situated as the following image shows.

The T-coffee alignment was a little insufficient for the proteins’ length because some amino acids were missing at the beginning and at the end of the predicted protein. It also didn’t have a Met at the beginning of the sequence. However, different cysteine residues could be found in both compared sequences.
No prediction of the protein and or SECIS element could be obtained with Seblastian, which indicates that there were no SECIS elements in this protein.
With all this information we reckon that PSTK from Callopanchax toddi is not a selenoprotein because it doesn’t have any selenocysteine residue nor SECIS element, so it’s a cysteine homolog.
SBP2
13 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000628_2.0). 10 belonged to scaffold SSNR01000084.1 (84.1) and 3 to SSNR01000122.1 (122.1)
In order to clarify which hit belonged to each reference sequence, a phylogenetic tree was built with the predicted sequences for all scaffolds of SBP2. The tree obtained can be seen below.

Scaffold 122.1 was chosen as closest to the query and Seblastian was performed. It was located between positions 65310 and 106291 in the positive strand. It had 6 exons, that were situated as the following image shows.

The T-coffee alignment was poor because the homology was insufficient for the proteins’ length. Due to the fact that SBPS is not a selenoprotein, a selenocysteine residue couldn’t be found in neither compared sequences.
Two SECIS structures were obtained but a protein prediction wasn’t when using Seblastian. Only one of them was located in the positive strand and in positions 76011-76101 from 3'-UTR region, 30280 nucleotides before the end of the gene. So, none of them could be a valid candidate.
All the information gathered confirms that SBP2 from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue nor a SECIS element. It is very similar to the Zebrafish homologue, so we could argue that they are orthologous genes.
SECp43
10 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000665_2.0). 3 belonged to scaffold SSNR01000042.1 (42.1), 5 to scaffold SSNR01000437.1 (437.1) and 2 to SSNR01000138.1 (138.1)
In order to clarify which hit belonged to each reference sequence, a phylogenetic tree was built with the predicted sequences for all scaffolds of SECp43. The tree obtained can be seen below, previously removing different predictions because the sequences were too short and gave worse T-coffee alignments than the others.

Scaffold 437.1 was chosen as closest to the query and Seblastian was performed. It was located between positions 204272 and 199173 in the negative strand. It had 8 exons, that were situated as the following image shows.

The T-coffee alignment was valid because the homology was sufficient for the proteins’ length, even though some gaps were present. Due to the fact that SECp43 is not a selenoprotein, a selenocysteine residue couldn’t be found in neither of the compared sequences.
One SECIS element was obtained but a protein prediction wasn’t when using Seblastian. It was located in the positive strand, so it couldn’t be a valid candidate.
All the information gathered confirms that SECp43 from Callopanchax toddi is a cysteine homologue because it doesn’t have a selenocysteine residue nor a SECIS element. It is very similar to the Zebrafish homolog, so we could argue that they are orthologous genes.
SecS
In this case, 9 hits were detected when aligning with the analogous protein from Danio rerio (SPP00000065_2.0). All of them belonged to scaffold SSNR01000145.1 (145.1).
The prediction found in this scaffold was located between positions 175590 and 155750 in the negative strand. It had 11 exons, that were situated as the following image shows.

The T-coffee alignment was valid for the proteins’ length because the homology was sufficient for the proteins’ length, even though some amino acids were missing at the end of the predicted protein. It also had a Met at the beginning of the sequence. However, a selenocysteine residue couldn’t be found in the compared sequences, due to the fact that it was a cysteine homologue.
One SECIS element was obtained but a protein prediction wasn’t when using Seblastian. It was located in the positive strand, so it couldn’t be a valid candidate.
With all this information we reckon that SecS from Callopanchax toddi is not a selenoprotein because it doesn’t have any selenocysteine residue nor SECIS element.
eEFsec
8 hits were detected when aligning with the analogous protein from Danio rerio (tr|F1R2J6|F1R2J6_DANRE). 7 belonged to scaffold SSNR01000078.1 (78.1) and 1 to SSNR01000033.1 (33.1)
A phylogenetic tree didn’t need to be built in this case because the T-coffee prediction of the scaffold 33.1 was empty. For this reason scaffold 78.1 was chosen as closest to the query. It was located between positions 2284623 and 2271858 in the negative strand. It had 7 exons, that were situated as the following image shows.

The T-coffee alignment was valid for the proteins’ length because the homology was sufficient for the proteins’ length. However, a selenocysteine residue couldn’t be found in the compared sequences because they were cysteine homologues.
No prediction of the protein or SECIS element could be obtained with Seblastian, which indicated that there was no SECIS element in this protein.
With all this information we reckon that eEFsec from Callopanchax toddi is not a selenoprotein because it doesn’t have any selenocysteine residue nor SECIS element, and is a cysteine homologue.