Introduction
Selenium
Selenium (Se) is the 34th element on the periodic table. It is a nonmetal that can be found in inorganic and organic forms in Earth. In its inorganic form it can be found as part of several mineral compounds such as selenite and selenide. In its organic form it can be found in the amino acids selenomethionine, selenocysteine, and methylselenocysteine. It also acts as a cofactor in some redox reactions, such as those in glutathion peroxidases. It is considered a macronutrient, and a lack of selenium can cause complications such as Keshan disease[2,3]
Selenoproteins
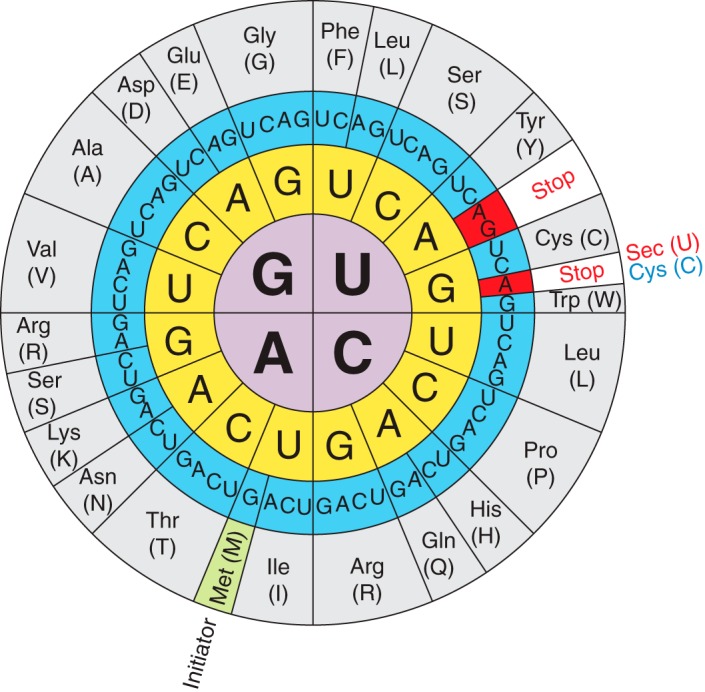
Figure 1.The genetic code illustrating the dual function of the UGA codon and that Sec is the 21st amino acid that is encoded by UGA.
Selenoproteins are families of proteins which contain selenocysteine. They have been found in every domain of life, but in eukaryotes they show a heterogeneous distribution. For example, vertebrates and algae have dozens of these proteins, while other organisms, such as higher plants and fungi, have lost all selenoproteins during evolution or their Secs have been replaced by Cys. Also, bony fishes are characterized by duplications of several selenoprotein families. [2]
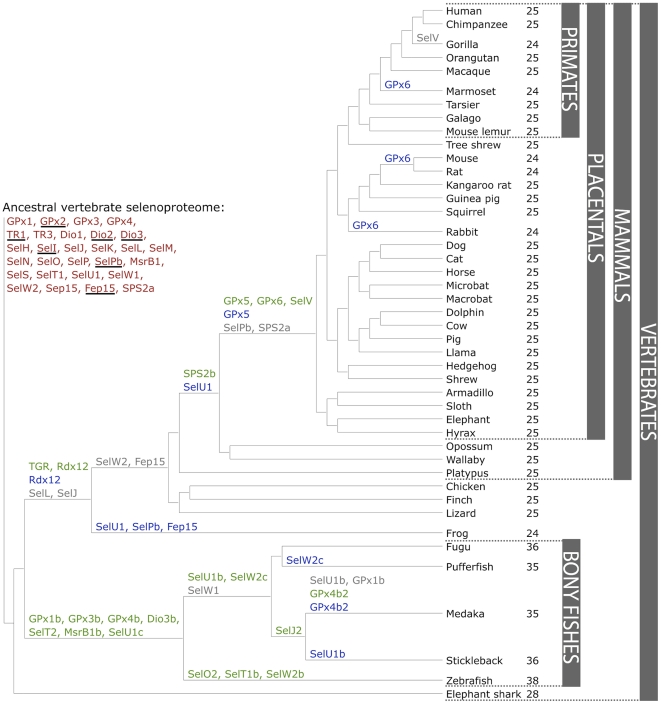
Figure 5. Evolution of the vertebrate selenoproteome.(Mariotti M et al.,2012)
Selenocysteine (Sec or U) is a nonstandard aminoacid, the use of which requires special machinery in order to be transcribed. It is an analogue of cysteine where the thiol group has been substituted for a selenol (HSe) group. The codon for selenocysteine is UGA, which usually acts as a stop codon. This is why additional translational machinery is needed for this UGA to be interpreted as a selenocysteine instead of a stop. [2,3]
Biosynthesis of Selenoproteins
In eukaryotes, selenoprotein translation is elicited by a set of dedicated cis-acting factors (SECIS elements and the in-frame UGA codon) and trans-acting factors (SPS1, SPS2, SecS, PSTK, eEFsec, SBP2 and the Sec-tRNA[Ser]Sec). This provide the process with a high grade of translational control
The tRNA needed to tranlsate UGA codon into a Sec amino acid has to suffer some undergo some biochemical modifications. It is first aminoacylated with Serine, then phosphorylated to form phosposeryl-tRNA[Ser]Sec by phosphoseryl-tRNA[Ser]Sec kinase (PSTK) and converted to selenocysteyl-tRNA[Ser]Sec (Sec-tRNA) by Sec synthase (SecS), the enzyme that incorporates the selenium in a monoselenophosphate form, which has been obtained from the dietary selelenium by selenophospate synthetases (SPS).
In order to incorporate the Sec-tRNA into the peptide under translation, some additional proteins are needed. SECIS binding protein 2 (SBP2) binds to the SECIs element from the mRNA via due to a RNA-binding domain that is known to bind SECIS elements with high affinity and specificity. SBP2 also interacts with the Sec-specific translation elongation factor (eEFSec), which recruits Sec-tRNA and facilitates incorporation of Sec into growing polypeptide (348). Furthermore, since the discovery of SBP2, additional SECIS-binding proteins were identified and their roles in selenoprotein synthesis were characterized, including ribosomal protein L30, and nucleolin, which are not essential but they can be involved in regulation processes in this context. [2,3,4]
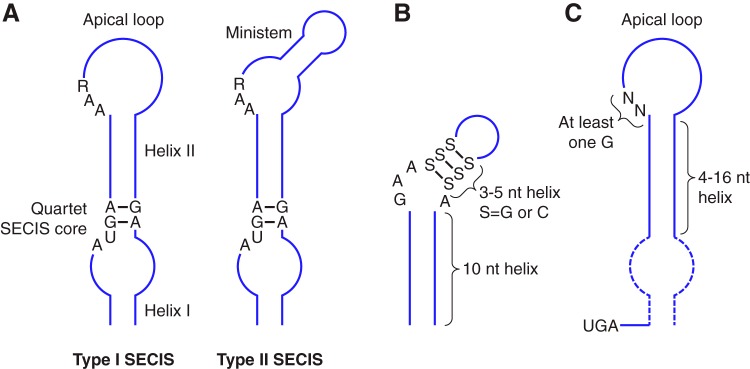
Figure 2 |
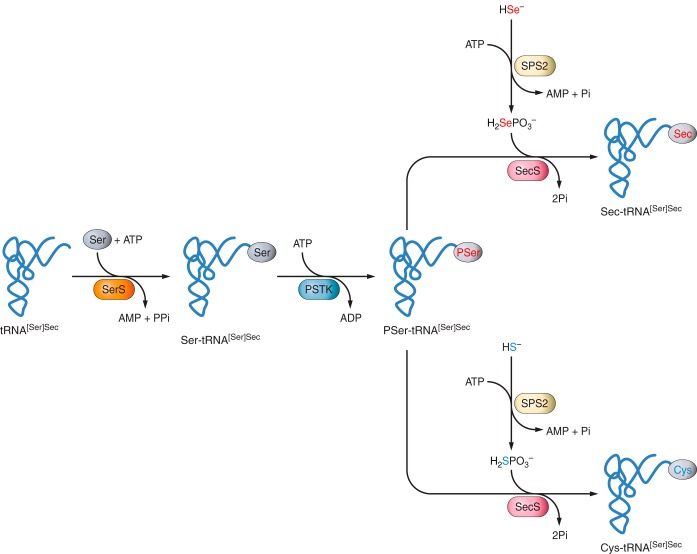
Figure 3 |

Figure 4 |
-->
|---|
Known Selenoproteins
The following list corresponds to the selenoproteins found in Danio rerio, a close relative of Danionella dracula and the organism against which we compare it.
- 15 kDa selenoprotein (Sel15): Sep15 has a thioldisulfide oxidoreductase function, and seems to be related to quality control for correct protein folding. Downregulation of seel15 has been related to cancer, especially malignant mesotheliomas.
- Fish selenoprotein 15 (SELENOE): It is an ER protein with an unknown function, occuring only in fish.
Glutathione peroxidase (GPx) family
- Glutathione peroxidase 1a and 1b (GPx1a-1b): Part of the GPx family, 1a and 1b, they are expressed in all cell types and serve as a reductor agent to turn H2O2 to H2O
- Glutathione peroxidase 2 (GPx2): Mainly expressed in the digestive tracts of organisms, especially the floor of the intestinal crypts. It acts as a protector against oxidative stress in the intestines, and it is capable to reduce a wider variety of substrates (mainly peroxidases) than GPx1. It appears to upregulate in gastrointestinal cancers, which leads to believe it has protective function against tumors.
- Glutathione peroxidase 3 a and b (GPx3a-b): A secreted version of GPx found in plasma and extracellular matrix, probably protecting extracellular tissues from oxidative damage.
- Glutathione peroxidase 4 a and b (Gpx4a-b): An ubiquitously expressed protein, it has a protective function against oxidation of lipid peroxides. It appears to be essential for development in vertebrates, and specially crucial to rain development and maintenance during maturity. Unlike other proteins of the GPx family, GPx4 has a structural function as well as an enzymatic one, as it becomes part of the mitochondrial sheath in spermatozoa.
- Glutathione peroxidase 7: Another peroxidase with ubiquitous expression, with a marked overexpression in tumoral tissues
- Glutathione peroxidase 8: Besides the enzymatic activity common to the GPx family, Gpx8 takes part in regulation of calcium fluxes in the cytosol and mitochondria.
- Methionine sulfoxide reductase A (MsrA): This enzyme, specifically expressed in kidney and nervous system, reduces methionine sulfoxide to methionine, repairing oxidative damage to proteins.
- Selenoprotein H (SELENOH): A nuclear protein, it appears to regulate expression of genes related to de novo glutathione synthesis.
- Selenoprotein I (SELENOI): With ubiquitous expression, this protein had no know function until recent studies have found that it might be part of a phospolipid synthesis pathway common to most tissues.
- Selenoprotein J (SELENOJ): A selenoprotein found only in certain fish, it has a structural function by providing transparency and correct light refraction to eye lenses.
- Selenoprotein K (SELENOK): This protein is located in the membrane of the endoplasmic reticulum, and altough it seems to protect against oxidative damage, its exact function is unclear.
- Selenoprotein L (SELENOL): The structure of selenoprotein L, presenting thioredoxin-like folds (UxxU, where U is a selenocysteine), suggest it has a redox function. Its distribution is limited to a few species of aquatic eukaryotes, Zebrafish being one of them.
- Selenoprotein M (SELENOM): This protein is similar to Sel15, and it also presents an oxidoreductase function.
- Selenoprotein N (SELENON): SELENON is a glycoprotein present in the endoplasmic reticulum, and it has antioxidative and calcium homeostasis functions.
- Selenoprotein O (SELENOO): Selenoprotein O is one of the few selenoproteins with a still unknown function, even though it has homologues in a wide variety of animal species.
- Selenoprotein P (SELENOP): The only selenoprotein with more than one Sec residue (17 in zebrafish), selenoprotein P is found in plasma and accounts for most presence of plasmatic selenium. It functions as an extracellular antioxidant.
- Selenoprotein R (MSRB): Distributed through all tissues but specially in liver and kidneys, Selenoprotein R mitigates oxidative damage to methionine residues in proteins
- Selenoprotein S (SELENOS): Expressed in the membrane of the endoplasmic reticulum, this protein takes part in the degradation process of misfolded proteins in the ER.
- Selenoproteins T, U, W (SELENOT, SELENOU, SELENOW): These three proteins have high levels of sequence homology, and even though their precise function is not known, the presence of thioredoxin folds in their structures suggest a redox function
- Thioredoxin reductase (TXNRD): This enzyme acts as an oxidoreductase , using NADH to reduce oxidised thioredoxin.
Iodothyronine deiodinase family
- Iodothyronine deiodinase 1 (DIO1): This membranal enzyme is dedicated to the conversion of thyroid prohormone T4 into T3.
- Iodothyronine deiodinase 1(DIO2): Like DIO1,DIO2 aids in the conversion if T4 to T3, but while DIO1 does this conversion in the thyroid gland and peripheral tissues, DIo2 generates it locally in brown adipose tissue, pituitary gland and the brain (ref. gran)
- Iodothyronine deiodinase 3 a and b (DIO3a-b): These membrane-associated proteins act as a deactivator of the Thyroid hormone.[2,3, 6, 7,11, 12]
Known Machinery
- Eukaryotic elongation factor (eEFsec): Part of the selenoprotein machinery, it serves as an elongation factor for sec-trna when the UGA codon has to be interpreted as a SeC
- Phosphoseryl-tRNA kinase (PSTK): This protein is highly conserved in all species, and it is speculated to have an important role in selenoprotein synthesis and regulation.
- SECIS binding protein 2 (SBP2): Part of the selenoprotein machinery, as its name indicates this protein helps binding SECIS elements for traduction of selenoproteins.
- Selenocysteine syntase (SecS): This enzime acts in one of the steps of Selenocystein synthesis, the conversion of O-phosphoseryl-tRNA to selenocysteinyl-tRNA.
- Selenophosphate synthetase (SEPHS): SEPHS syntethises selenophospate using selenide and ATP as substrates.
- tRNA Sec 1 associated protein 1 (SECp43): This protein is associated with biosynthesis and incorporation of Sec, but its exact function is currently unknown. [2,3,6,7,12,13]
Danionella dracula
Origin and name
Danionella dracula is a freshwater fish endemic to Myanmar. The generic name Danionella is given to a genus of fishes belonging to the Danionidae subfamily, which in turn receives its name from the Bengali Dhani, meaning “from the paddy”, as these species usually inhabit rice paddies and other stillwater habitats. The name of the particular species Dracula comes from the characteristic fang-like protuberances in its upper jaw, resembling those of Bram Stoker’s character in the novel Dracula.[1]
Danionella dracula is amongst the smallest species of fish, with a maximum known length of 16,7 mm. It barely shows any pigmentation, sporting a simple dark band along its body, the rest of it being transparent. While relatively similar to other fishes of the Danionidae subfamily, a few particularities separate this species from the rest. The main

Figure 3. Danionella dracula by Peter McGuire
Distribution and habitat
Danionella dracula was found in a small stream near the town of Mongkawn in Myanmar. At the moment it has only been found in this location. In captivity, it has been shown to survive in temperate fresh water, much like the rest of cypriniforms.
Diet
Although its diet in the wild has not been documented, Danionella dracula has shown to feed on small crustaceans and invertebrates.
Phylogeny
The taxonomy of oyr organism is the following:
Kingdom: Animalia
Phylum: Chordata
Class: Actinopterygii
Order: Cypriniformes
Family: Cyprinidae
Subfamily: Danionidae
Genus: Danionella
Specie:Danionella dracula[1,8]
Click here for more information.


