In higher eukaryotes gens are interrupted by introns (non-coding sequences). They must be excised from pre-mRNA molecules to yield mature, functional mRNAs. It has long been known that intron removal and the ligation of flanking sequences (exons) occurs through two sequential trans-esterification reactions that are carried out by a multicomponent complex that is known as the spliceosome.
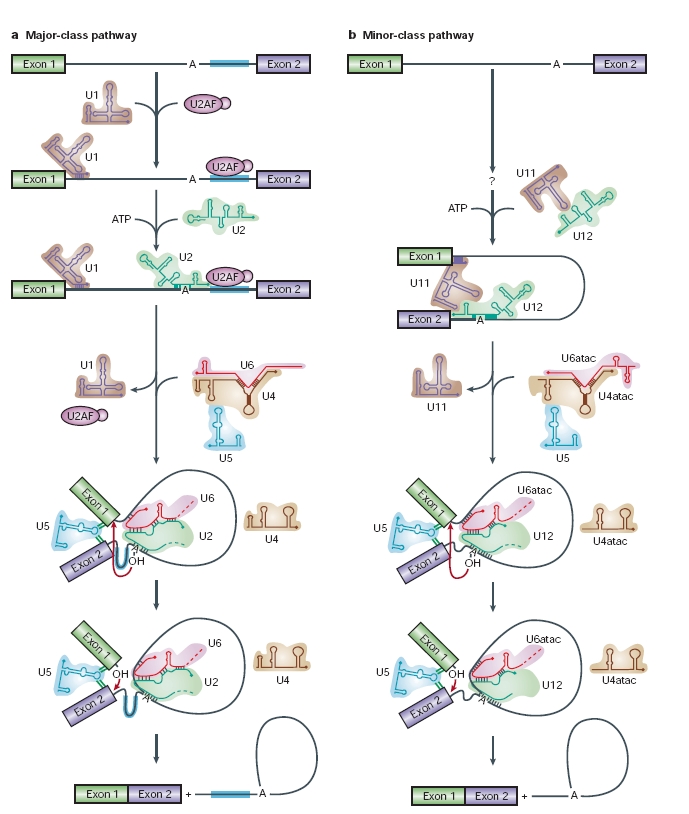
Most introns have common consensus sequences near their 5' and 3' ends that are recognized by spliceosomal components. The assembly of a spliceosome onto a pre-mRNA is a process that involves five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4, U5 and U6), as well as an array of protein factors. Catalysis of the splicing reaction leads to exon ligation.
Recently, metazoan genes have been found to contain a new class of rare introns that have non-canonical consensus sequences and are excised by a distinct splicing machinery. The low-abundance spliceosome that is responsible for the excision of these introns includes four snRNPs (U11, U12, U4atac and U6atac) that are different from, but functionally analogous to, the well-characterized U1, U2, U4 and U6 snRNPs, respectively. U5 snRNP is shared by both spliceosomes. Because the excision of these minor-class introns is dependent on the U12 snRNP, they are referred to as U12-type introns, whereas major-class introns that require the analogous U2 snRNP are referred to as U2-type introns. Although there are few U12-type introns in the genome of any given species, their persistence throughout virtually all of metazoan evolution indicates that they have an ancient origin and an important cellular function. The two spliceosomes have remarkable mechanistic similarities that highlight their common ancestry and enhance our understanding of the inner workings of the spliceosome.
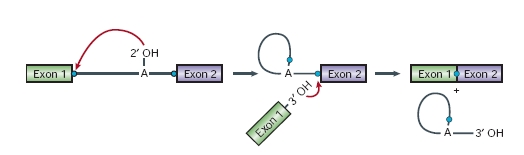
Splicing mechanismAs we have said, the spliceosome is a dynamic machine that catalyses two trans-esterification reactions:
In the first step, cleavage of the 5' exon-intron junction occurs on nucleophilic attack by the 2' hydroxyl group of a conserved adenosine residue at the intron branch site, upstream of the 3' splice site. This generates a free 3'hydroxyl group on the upstream exon, as well as a branched lariat intermediate. In the second step, the 3' intronexon junction is attacked by the 3' hydroxyl of the 5' exon, displacing a lariat intron and ligating the exons. After the second step has been completed, the ligated exons and a lariat intron are released, and the spliceosomal components dissociate and are recycled for further rounds of splicing.
U12 introns
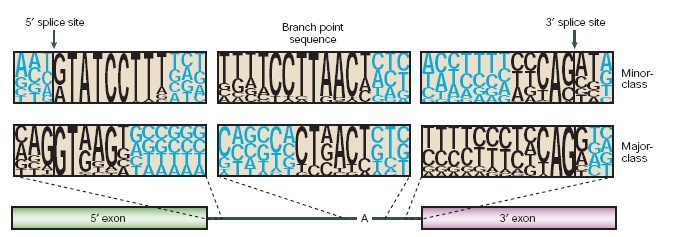
The first intron sequences ever characterized revealed highly conserved dinucleotides at the 5'and 3' termini (GT and AG, respectively), which were later found to be parts of longer consensus sequences at the 5'splice site and the 3'splice site. The presence of nonconsensus splice sites was recognized in 1994 when Hall and Padgett proposed that there was a distinct minor class of introns. They saw that four introns shared unusual consensus sequences, and predicted that their excision was mediated by a distinct spliceosome that involved low-abundance snRNPs.
These new introns had AT and AC termini, which deviates from the nearly invariant GT-AG rule. For this reason they were named them AT-AC introns. However, AT-AC termini are not a defining feature of minor-class introns. Most minor-class introns have canonical GT-AG termini, and, very rarely, major-class introns have AT-AC termini. U12-dependent splicing is determined by the longer and more tightly constrained consensus sequences at the 5'splice site and branch site of minor-class introns, as well as by the lack of a polypyrimidine tract upstream of the 3'splice site.
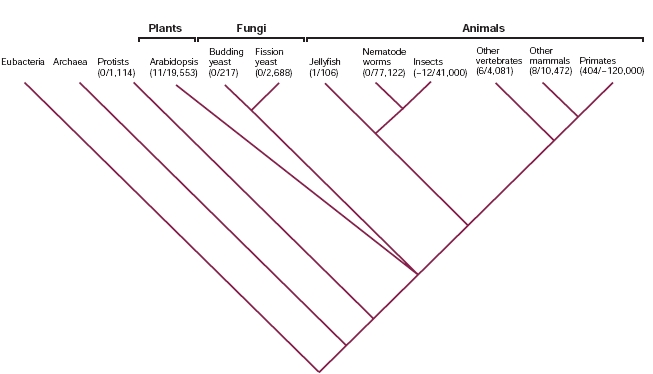
Examples of U12-type introns are found in plants and most of the metazoan taxa that have been examined (vertebrates, insects and cnidarians as jellyfish). There is no evidence of U12 introns, or of the U12-type spliceosome, in simple eukaryotes such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans and protists.
It has been estimated that the frequency of occurrence of U12-type introns is in the range of 0.15-0.34% relative to U2-type introns in vertebrates, but is lower in other metazoan taxa. In the image below we can see the frequency of occurrence of U12 introns in different taxa. Below each taxon, the number of U12 introns identified from the total number of introns analysed is indicated in brackets:
U12-type introns almost always coexist with neighbouring U2-type introns in a host gene, and do not show any positional bias relative to other introns in the gene. Although U12-type introns most often occur singly in any given gene, there are several genes that have two or more. The length of U12-type introns has a similar distribution to that of U2-type introns in humans (U12: mean = 3,600 bp; U2: mean = 4,130 bp).
U12-type introns are not restricted to a particular functional class of genes, although a large proportion of the genes function in the processing of genetic information, such as DNA replication and repair, transcription, RNA processing and translation. Few U12-type intron-containing genes are involved in metabolism or small-molecule biosynthesis.
 Click to enlarge!
Click to enlarge!