Selenium (Se) is a essential micronutrient for a vast number of species. When consumed in large doses, selenium is toxic for the organism, however, low doses of this element are important to carry out several physiological processes. Moreover, selenium is a component of the unusual amino acid selenocysteine[1,2].
Selenocysteine (Sec/U) is the 21st amino acid in the genetic code. It has a structure similar to that of cysteine, but it has selenium atom instead of a sulfur, which forms a selenol group.
Selenocysteine is encoded by the codon UGA, even though this is usually a termination codon. However, several organisms use this codon to incorporate selenocysteine into proteins called selenoproteins. Due to the fact that UGA normally encodes for a cessation of translation, organisms that synthesize selenoproteins require specific translation machinery[1,2,3,4,5].
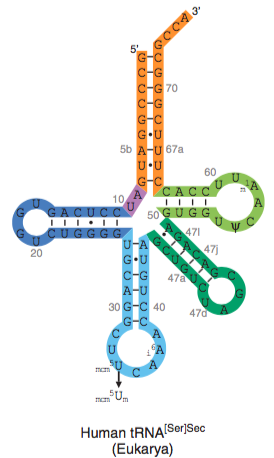
Biosynthesis of Sec greatly differs from other amino acids’. Sec is the only amino acid in eukaryotes whose biosynthesis occurs on its own tRNA, named Sec tRNA[Ser]Sec.
The first step for Sec biosynthesis is the transcription of its tRNA, which is the longest tRNA sequenced and forms unusual secondary structures.
The gene for tRNA[Ser]Sec is Trsp. Trsp is a single copy gene in most organisms except for fish and it encodes for the selenoprotein synthesis machinery. Its transcription is regulated by three upstream regions: a TATA box motif (30nt), a proximal sequence element (70nt) and a distal sequence element (200nt).
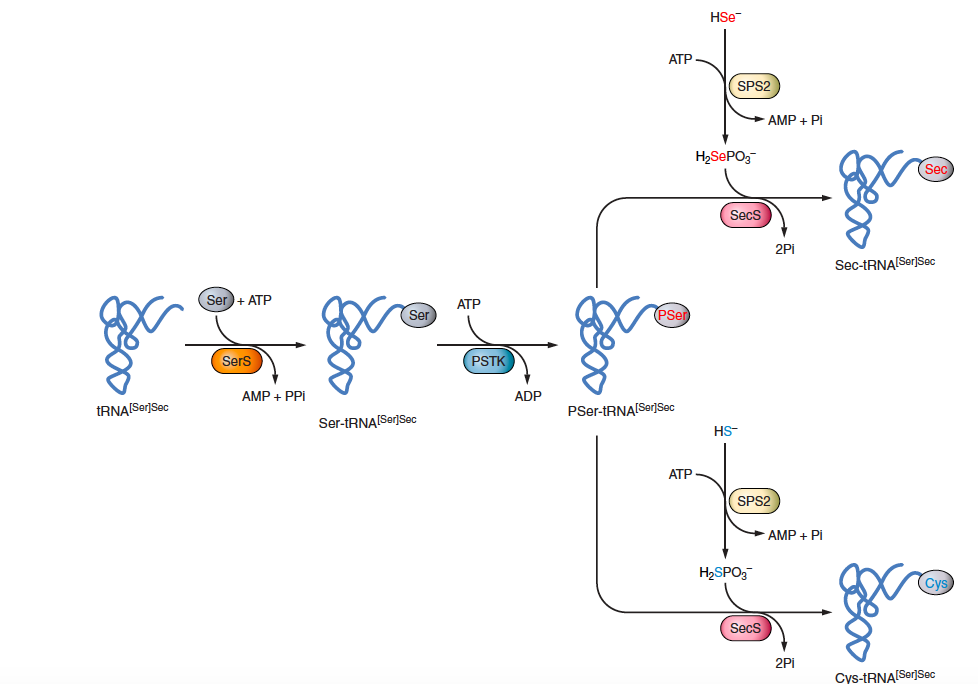
Once tRNA [Ser]Sec is transcribed, it is aminoacylated with Serine (Ser) by the enzyme Seryl-tRNA synthetase (SerS), yielding to Ser tRNA [Ser]Sec. Ser tRNA [Ser]Sec is then phosphorylated by Phosphoseryl tRNA Kinase (PSTK), giving rise to PSer tRNA [Ser]Sec. The last step is the conversion of the Ser moiety on PSer tRNA [Ser]Sec to selenocysteyl-Sec tRNA [Ser]Sec. This reaction is carried out by Sec synthase (SecS), which incorporates the active form of Se, selenophosphate, into the amino acid backbone (Figure 2).
Sec is degraded by the enzyme Sec lyase (SCL), decomposing Sec into L-alanine and elemental Se. Although the physiological role of SCL has not been yet discovered, some studies suggest its possible role in recycling the Se of Sec during degradation of selenoproteins.
Inorporation of Sec into proteins takes place during translation and it is dictated by UGA codons present in selenoproteins mRNA. The complex mechanism by which Sec is incorporated into proteins requires special trans-acting protein factors, Sec tRNA[Ser]Sec and a cis-acting Sec insertion sequence (SECIS) element.
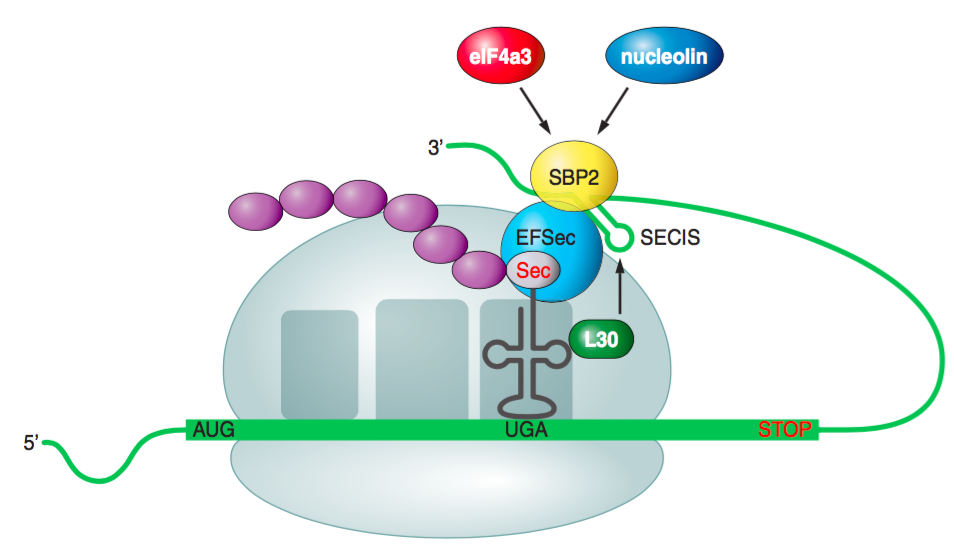
SECIS elements act as factors that guide the coding of UGA codon as Sec. When there is a SECIS element in an mRNA, Sec tRNA[Ser]Sec is able to translate UGA as a Sec. The recoding of Sec needs at least two trans-acting factors: SECIS binding protein 2 (SBP2) and Sec-specific translation elongation factor (eEFSec). Not only SBP2 interacts with ribosomes and SECIS elements, but also with eEFSec, so Sec tRNA[Ser]Sec is recruited and it enables the incorporation of Sec into new proteins.
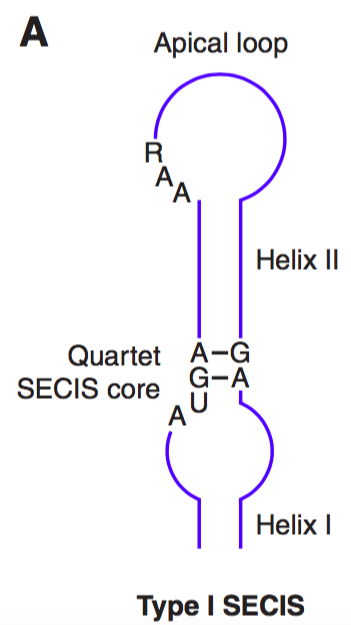
SECIS elements are cis-acting RNA structures found in the 3’UTR region of selenoprotein mRNA, immediately downstream of the UGA that encodes for Sec. SECIS elements are constituted by two helixes separated by an internal loop, a GA Quartet composed of 4 non-Watson.Crick interactions, an apical loop. The GA Quartet is the core of the SECIS element and it is needed for interaction with SBP2. Moreover, the apical loop has a conserved AAR motif required for Sec incorporation. Even though the function of the AAR motif is unknown and no AAR binding proteins have been found, it seems that this motif plays a role in the ribosome.
SECIS binding protein 2 (SBP2) is a limiting factor for selenoprotein synthesis. Studies in which SBP2 was knocked down, showed decreased expression of selenoproteins. SBP2 contains three domains: an NH2-terminal domain, a Sec incorporation domain (SID) and a COOH-terminal RNA binding domain (RBD). The role of the NH2-terminal domain is not known, but previous studies determined that it is not required for Sec incorporation, so it may have a regulatory function. On the other hand, RBD and SID form a complex that binds to the SECIS element. The RBD part id the one that interacts with the SECIS, while SID enhances that interaction.
The Sec-specific eukaryotic translation elongation factor (eEFSec) recruits tRNA[Ser]Sec and inserts Sec to the forming proteins in response to the UGA codon. It has a GTPase activity and high specificity for aminoacylated tRNA[Ser]Sec. It has four domains, one of them is involved in interactions with SBP2 and tRNA[Ser]Sec. In order to recruit the tRNA
L30 is a component of the large ribosomal subunit in eukaryotes. It binds to SECIS elements through an L7Ae RNA-binding motif that is also found in SBP2. L30’s function has not been fully studied but it may compose a part of the Sec insertion machinery.
Selenoproteins are proteins that contain Sec residues in their sequence. Usually, Sec is located in enzyme active sites so its main function is to perform catalytic redox reactions. Therefore, mutations of Sec to any other amino acid leads to enzyme inactivation.
At present, there are more than 50 selenoprotein families described. Their distribution differs notably among species, thus, analyses of selenoproteomes are important to study the evolutionary trends regarding the use of Sec. For example, some terrestrial animals have replaced selenoproteins for Cys homologs (losing selenoproteins), whereas most aquatic animals have kept their selenoproteomes. This finding indicates that the environment plays a role in the evolution of selenoproteins.[7]
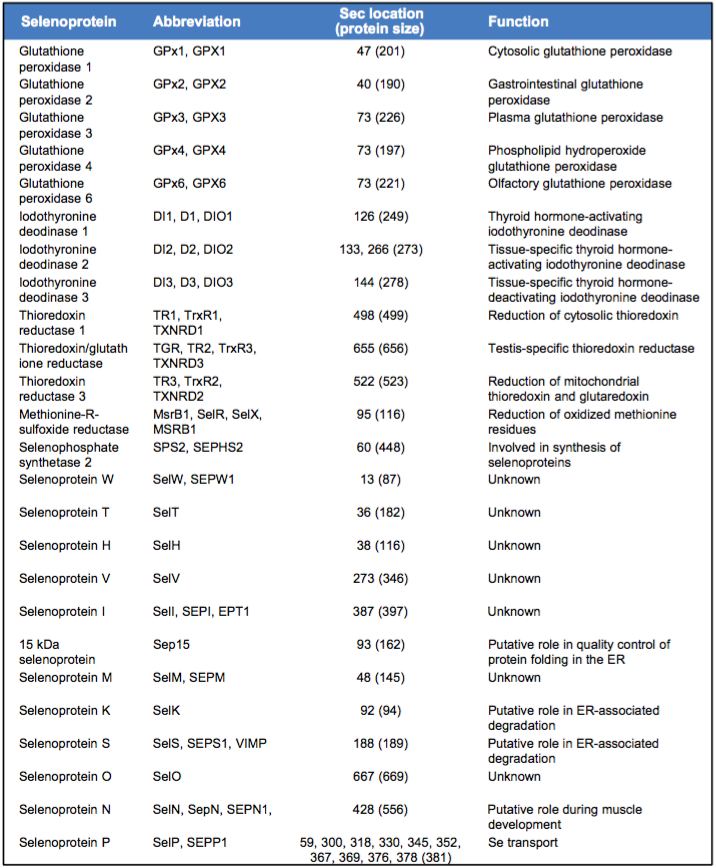
Nowadays, 25 families of selenoproteins in eukaryotes have been characterized. This table summarizes the Human Selenoproteome.
Since UGA has a dual function of both encoding for selenocysteine and cessating the translation process, there can sometimes be a missanotation in the database. That’s why the role of bioinformatics in this field is unquestionable and essential. There are two major bioinformatics approaches for identification of selenoproteins: the first one consists on finding the candidate SECIS elements in completely sequenced genomes so the UGA codon can be located upstream. However this approach does not account for different SECIS elements patterns across species. The second approach involves the identification of in-frame UGA codons in completely sequenced genomes, independently of SECIS elements.
In this study we use both approaches to identificate and characterize the selenoproteome of a recently sequenced species, Anarrhychtys Ocellatus[8].