
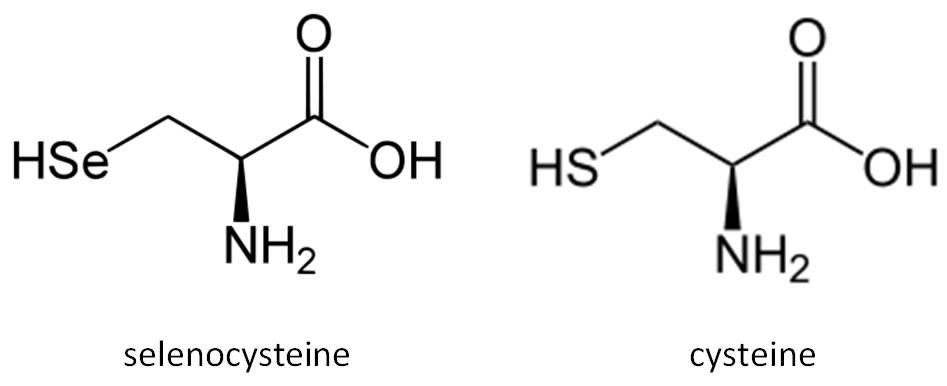
Selenium (Se) was discovered in 1817 by the Swedish chemist Berzelius. Today, selenium is well established as an essential trace mineral for which both beneficial and toxic effects in human health have been described. Selenium is an essential micronutrient of major metabolic significance. It is now clear that the importance of having adequate amounts of this micronutrient in the diet is primarily due to the fact that selenium is required for the biosynthesis of selenocysteine (Sec), the twenty first naturally ocurring amino acid in proteins. Selenium is known primarily for its antioxidant activity and, in therapeutic aspects, for its chemopreventive, antiinflammatory and antiviral properties.
Selenium becomes cotranslationally incorporated into the polypeptide chain as part of the amino acid Sec. It is a rare amino acid in extant proteins that is chemically similar to cysteine (Cys). The remarkable aspect of Sec incorporation into proteins is that it is encoded by the UGA codon, which in most circumstances signals translation termination. This codon duality is circumvented by the presence of conserved cis- and trans- acting elements and protein factors dedicated to the decoding of UGA as Sec (Allmang et al, 2006; Lobanov et al, 2009; Castellano et al, 2009).
The proteins that contain a selenocysteine as an integral part of their polypeptide chain are defined as selenoproteins. Most selenoproteins contain a single selenocysteine residue per polypeptide chain. However, there are exceptions like SelP, which contains a lot of selenocysteines (Talat et al, 2000). Because Sec is encoded by one of the stop codons, selenoproteins are difficult to identify in the genome and are generally misannotated (Mariotti et al, 2010). However, selenoproteins are present in the three domains of life: bacteria, archaea and eukaryota. Several selenoprotein families have been identified. Selenoproteomes among species are generally small; the largest repertoire exists in fish. Humans and rodents have 25 and 24 selenoproteins, respectively. Their small size can be explained by a limited selenium supply in nature and a rather energy-expensive synthesis process. For most selenoprotein families, Sec/Cys interconversion is commonly observed across species (Vanda et al, 2007).
The human selenoproteome consists of 17 selenoprotein families, some with multiple genes with similar functions. These include glutathione peroxidases (GPx), thioredoxin reductases (TrxR), iodothyronine deiodinases (DIO), and selenophosphate synthetases 2 (SPS2). The remaining selenoproteins have been annotated in alphabetic order and include Sel15, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR, SelS, SelT, SelV and SelW. Only a few of them have been functionally characterized, but the functions of most other selenoproteins remain unknown. Mapping the selenoproteomes in species across the domains has paved the way for the main challenge: the functional characterization of these proteins and their involvement in the etiology of disease.
Back to top
Translation of selenoproteins is similar to generalized protein translation in that it consists of the three main steps: initiation, elongation and termination. The special feature of selenoprotein translation lies in the recoding of the UGA codon, which is located in the coding region of selenoprotein mRNAs, from a stop codon to Sec-insertion codon (Talat et al, 2000).
Typically, the translational machinery reads the UGA codon as a termination signal, releasing the polypeptide from the ribosome. In the case of selenoproteins, cis- and trans-acting factors work to redirect the machinery to insert selenocysteine instead of terminating the synthesis. These are the basic elements needed for the biosynthesis of selenoproteins:
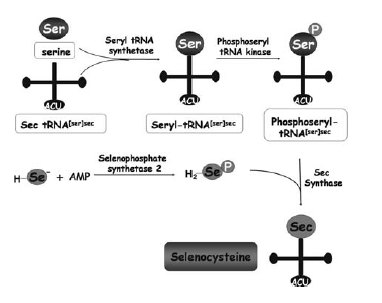
First, the tRNA carrying Sec (tRNA[Ser]Sec) needs to be synthesised. This synthesis shares some steps with the synthesis of the tRNA for cysteine. Briefly, seryl-tRNA synthase aminoacylates selenocysteyl-tRNA[Ser]Sec. The seryl moiety is then phosphorylated by phosphoseryl-tRNA[Ser]Sec kinase (Pstk). Then, Sec synthase (SecS) replaces the phosphoryl moiety of phosphoserine by selenide. This is derived from the selenium donor selenophosphate, which is synthesised by selenophosphate synthetase 2 (SPS2), a selenoprotein itself (Vanda et al, 2007). Selenophosphate synthetase 1 (SPS1) is a related protein that contains a Cys residue in place of Sec, but its role in selenoprotein synthesis or any other biological process is unknown. tRNA selenocysteine 1 associated protein 1 (Secp43) is also involved in the synthesis of the tRNA[Ser]Sec and selenoproteins (Reeves et al, 2009).
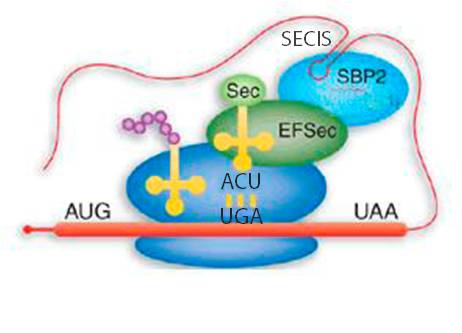
The main signal that is required by the ribosome in order to differentiate properly a stop codon from a selenocysteine codon is the SECIS (SElenoCysteine Insertion Sequence) element, which is a secondary structure element of the mRNA. It consists of a sequence around 60 nucleotides in length that adopts a stem-loop structure. This structural motif directs the cell to translate UGA codons as selenocysteines. In this way, SECIS elements are a fundamental aspect of messenger RNAs encoding selenoproteins. SECIS elements occur in the 3' UTR of the mRNA in archaea and eukaryota. This element recruits SECIS Binding Protein 2 (SBP2), which binds and recruits the selenocysteine specific elongation factor (EFsec). This protein selects specifically the tRNA[Ser]Sec, which is taken to the UGA codon of the mRNA. In this way, the selenocysteine incoporation is achieved. The translation of mRNA continues until the ribosome recognizes a new termination codon (Reeves et al, 2009; Talat et al, 2000; Vanda et al, 2007).
Now we present a litte explanation of each human selenoprotein or selenoprotein family. We expect that if we find homologous proteins in Saimiri boliviensis, they will conserve similar functions.
Glutathione Peroxidases
The glutathione peroxidase (GPx) family is one of the most fully characterized groups of selenoproteins. This family contains from GPx1 to GPx8. In humans, GPx1 through 4 and GPx6 are Sec-containing enzymes, whereas GPx5, GPx7 and GPx8 are cysteine-containing homologues. In vitro activity assays suggest that all members of this group use glutathione to catalyze the reduction of hydrogen peroxide and/or phospholipid peroxides, but the physiological localization and substrate specificity of each varies, collectively providing a wide spectrum of antioxidant protection (Reeves et al, 2009).
Thioredoxin Reductases
Thioredoxin reductase (TR) enzymes are oxidoreductases that use NADPH to catalyze the reduction of oxidized thioredoxin (Trx). Trx is in turn used by several cellular enzymes as a cofactor in dithiol-disulfide exchange reactions and this is a major mechanism by which a reduced environment is maintained within cells, particularly serving to maintain reduced cysteine groups. There are three mammalian TRs: cytoplasmic/nuclear TR1, mitochondrial TR2, and testes-specific thioredoxin-glutathione reductase TR3 (Reeves et al, 2009).
Deiodinases
The iodothyronine deiodinase family of selenoproteins consists of three enzymes: types 1, 2 and 3 (DI1, 2 and 3; or DIO1, 2 and 3), which are membrane-anchored enzymes of 29-33 kDa that share substantial sequence homology and catalytic properties. Thyroid hormone action is initiated by the activation of T4 prohormone to T3. This conversion is carried out by D1 or D2. T4 and T3 are irreversibly inactivated via inner ring monodeiodination catalyzed by D3. Thus, thyroid hormone metabolism is dependent upon the combined actions of the three deiodinases (Reeves et al, 2009).
Selenoprotein 15
Studies suggest that Selenoprotein 15 (Sel15) may have redox function and may be involved in the quality control of protein folding. This selenoprotein localizes to the ER. A link between Sel15 and cancer has been made by several in vitro studies (Reeves et al, 2009).
Selenoprotein H
Selenoprotein H (SelH) is a 14kDa thioredoxin fold-like protein that contains a conserved Cys-X-X-Sec motif. Its expression is widely distributed throughout a variety of tissues and relatively high in early stages of embryonic development. SelH was originally identified in Drosophila melanogaster, where it was found to be essential for viability and antioxidant defense. SelH is localized to the nucleus and overexpression studies suggest it functions in regulating expression levels of genes involved in de novo glutathione synthesis and phase II detoxification in response to redox status (Reeves et al, 2009).
Selenoprotein I
Selenoprotein I (SelI) has been found to contain sequence homology to enzymes involved in phospholipid synthesis. RT-PCR and Northern blot analysis revealed that human SelI was ubiquitously expressed in multiple tissues, consistent with the notion that it is involved in a phospholipid byosynthesis pathway common to most tissues (Reeves et al, 2009).
Selenoprotein K
Selenoprotein K (SelK) is a small (16kDa) protein localized to the endoplasmic reticulum membrane and some evidence suggests it is associated with the plasma membrane. Expression of SelK has been suggested to be relatively high in the human heart. However, more recent studies demonstrated that mRNA levels are widely distributed throughout mouse tissues, with particularly high levels detected in spleen and testes. The function of SelK remains unclear (Reeves et al, 2009).
Selenoprotein M
Selenoprotein M (SelM) has Cys-X-X-Sec motifs. This selenoprotein is 15kDa and shares 31% sequence identity with Sel15. Both of them localize to the ER. Different studies have suggested these two selenoproteins function as thiol-disulfide oxidoreductases. They have also been suggested to play a role in protein-folding in the ER (Reeves et al, 2009).
Selenoprotein N
Similar to SelK and SelS, selenoprotein N (SelN) is a transmembrane protein localized to the ER membrane, but much larger in size (70kDa). There are two known isoforms of the SelN gene product, with isoform 1 corresponding to the full-length transcript and isoform 2 excluding exon 3 from splicing. Several independent studies have linked mutations in SelN gene to muscular disorders (Reeves et al, 2009).
Selenoprotein O
Selenoprotein O (SelO) is one of the selenoproteins that has remained enigmatic since identification of its sequence in the human genome. The presence of a Cys-X-X-Sec motif is suggestive of a redox function (Reeves et al, 2009).
Selenoprotein P
Selenoprotein P (SelP) is a unique member of the selenoprotein family in that it contains multiple Sec residues per protein molecule. Specifically, both human and mouse SelP contain 10 Sec residues. SelP contains 40-50% of the total Se in plasma, suggesting this protein may act as a Se transporter. SelP has the capacity of acting like a phopholipid hydroperoxide glutathione peroxidase (Reeves et al, 2009).
Methionine sulfoxide reductase A (MsrA)
This protein is a cysteine homologue in mammals that is ubiquitous and highly conserved. It carries out the enzymatic reduction of methionine sulfoxide to methionine. Its proposed function is the repair of oxidative damage to proteins to restore biological activity. Three transcript variants encoding different isoforms have been found (NCBI).
Selenoprotein R
Selenoprotein R (SelR, MsrB1) is part of the methionine sulfoxide reductase (Msr) family of proteins, which also includes MsrA, SelR2 and SelR3. Reactive Oxygen Species can oxidize methionine residues in proteins to produce a mixture of S- and R-forms of methionine sulfoxide, which are reduced by MsrA and SelR enzymes, respectively. SelR1 is the only member of the Msr family that is a selenoprotein. The presence of Sec instead of Cys at the active site of SelR1 has been suggested to have certain catalytic advantages and disadvantages (Reeves et al, 2009).
Selenoprotein S
Selenoprotein S (SelS) is a transmembrane protein located in the ER and plasma membranes and is widely expressed in a variety of tissues. It has been suggested to participate in the removal of misfolded proteins from the ER lumen for degradation and to protect cells from oxidative damage and ER stress-induced apoptosis. Expression of SElS has been shown to be modulated by glucose metabolism and ER stress (Reeves et al, 2009).
Selenoprotein T
Selenoprotein T (SelT) is a member of a subfamily of selenoproteins (also including SelW, SelH and SelV) that share sequence similarity containing a thioredoxin-like fold and a conserved Cys-X-X-Sec motif. The expression of SelT is proposed to be similar to those selenoproteins involved in stress-related phenomena. A study has shown that SelT has a biological role in calcium mobilization (Reeves et al, 2009).
Selenoprotein U
Selenoprotein U (SelU) is a family of three proteins containing Cys instead of Sec in mammals (in other organisms like C. intestinalis it is a selenocysteine): SelU1, SelU2 and SelU3. The function is unknown.
Selenoprotein V
Expression of this selenoprotein appears to be restricted to testes, but its function in this tissue is unknown. SelV shares sequence homology with other selenoproteins, including SelH, SelT and SelW, and these selenoproteins all possess a thioredoxin-like fold including a conserved Cys-X-X-Sec motif, suggesting redox functions (Reeves et al, 2009).
Selenoprotein W
Selenoprotein W1 (SelW1) is a small selenoprotein that contains a Cys-XX-Sec motif. Levels of SelW1 mRNA are highly dependent on adequate dietary Se levels as well as levels of SelP. SelW1 interacts with glutathione and evidence suggests it plays an antioxidant role in cells. It has also been suggested that SelW1 functions in muscle growth and differentiation by protecting the developing myoblasts from oxidative stress (Reeves et al, 2009). It is a member of the selenoprotein W family, which also includes SelW2, a cysteine-containing protein in humans (SelenoDB).
Taxonomic classification:
Kingdom: Animalia
Phylum: Chordata
Class: Mammalia
Order: Primates
Suborder: Haplorrhini
Infraorder: Simiiformes
Family: Cebidae
Subfamily: Saimiriinae
Genus: Saimiri
Species: Saimiri boliviensis
Subspecies: Saimiri boliviensis boliviensis
Saimiri boliviensis (Bolivian squirrel monkey) is a South American monkey, found in Bolivia, Brazil and Peru. These monkeys are typically arboreal, residing in the canopy among the small branches. However, they will occasionally leave the canopy to the shrub layer or the forest floor to scavenge. They occur at altitudes from sea level to 1,500 meters.
Squirrel monkeys are all fairly similar in appearance. The head is elongated and egg-shaped. The fur of Bolivian squirrel monkeys is dense and short, and is generally a yellowish tan color, mottled with black hair tips. The fur on the undersides of the limbs is yellow, white or orange. Males and females are very similar in appearance, with sexual dimorphism ocurring in size and color of crown fur. One key identifying feature of Saimiri boliviensis that differs from other squirrel monkeys is the arched eyebrows.
Within a troop of Bolivian squirrel monkeys, mature males live in a subgroup, generally separate from a female/young subgroup. This level of segregation between males and females is unique among Bolivian squirrel monkeys.
The diet of Saimiri boliviensis consists primarily of insects and fruits. Other foods eaten include berries, nuts, flowers, seeds, etc. Having a primary diet of insects and fruits, Saimiri boliviensis would play several important roles in the ecosystem (Saimiri boliviensis information).
Saimiri boliviensis and Homo sapiens are predicted to have a common ancestor around 45 milion years ago (Chiou et al, 2011).
Back to top