FINAL DATA
1. Dataset 22SIN3set: 22 genes from SIN3set
2. Running MEME in the 22SIN3set
- MEME interesting motif
TTTAAATATTATAAAAAAAATATAAATATATAAAATTTATTATAATATTT
E-value = 8.0e-018
In 21 out of 22 sequences (CG12505, FBgn0033926) - Log matrix
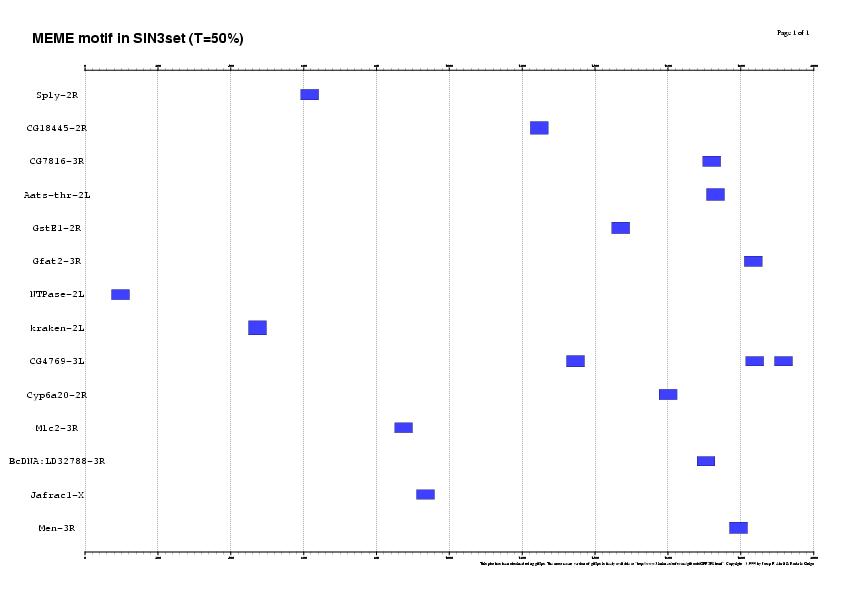
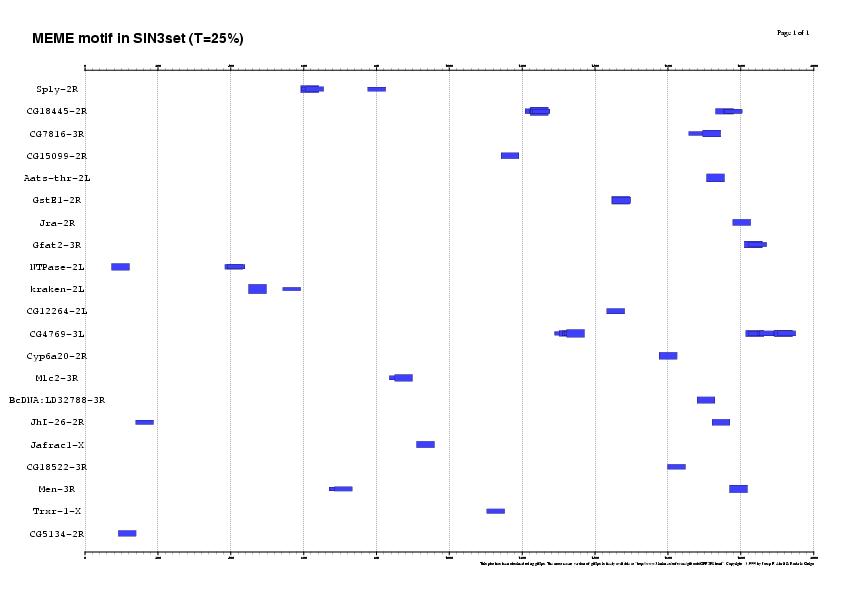
3. Running the matrix with different thresholds in 22SIN3set
Results sorted by matrix similarity score
MAXSCORE: 5136.0
| T = 50% | T = 25% | T = 0% |
|---|---|---|
 |
 |
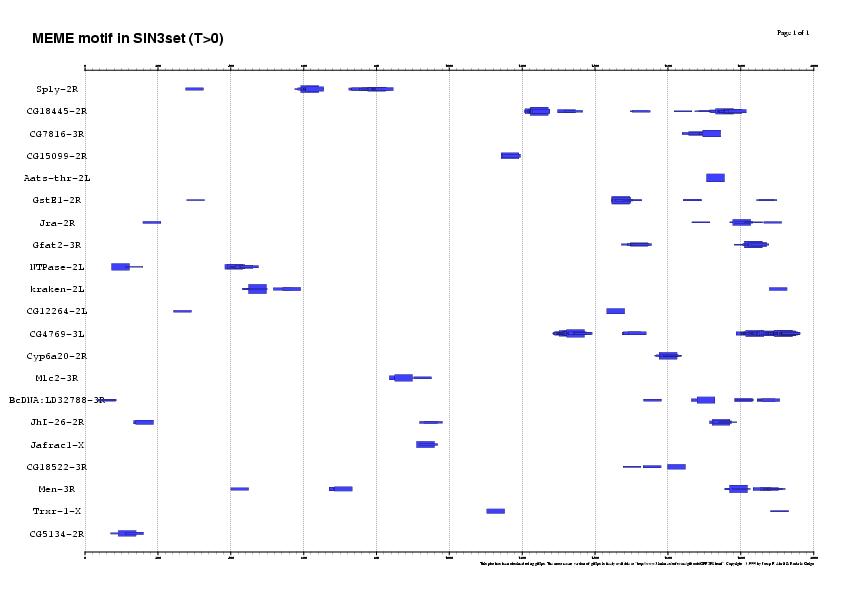 |
| 16 SITES (14 genes, 63%) | 53 SITES (21 genes, 95%) | 256 SITES (21 genes, 95%) |
| Genomic annotation |
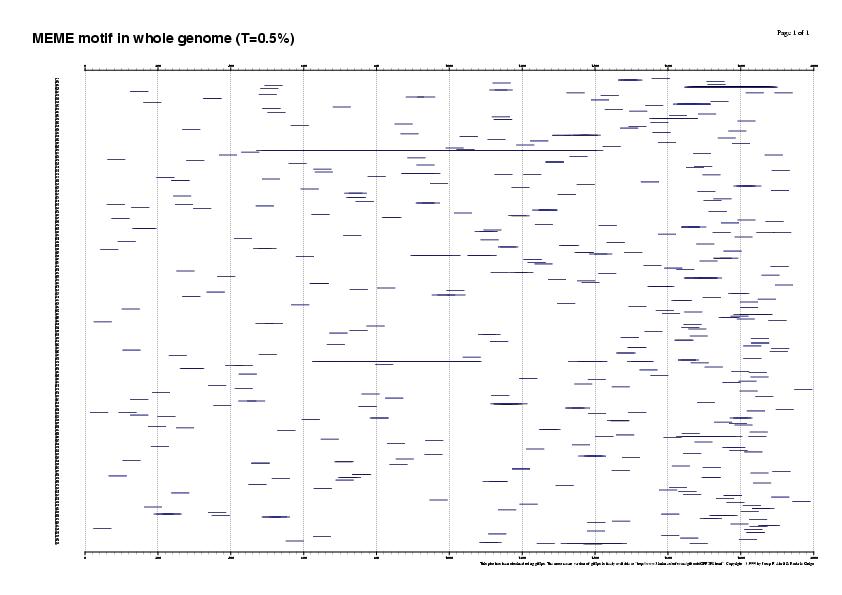
4. Running the matrix with different thresholds in the whole genome (13728 genes)
Results sorted by matrix similarity score
MAXSCORE: 5136.0
| T = 50% | T = 25% | T = 0% |
|---|---|---|
 |
 |
 |
| 862 SITES (330 genes, 2%) | 15386 SITES (4923 genes, 35%) | 115910 SITES (11590 genes, 84%) |
5. Genes whose putative sites (T=50%) are in positions [1500-2000]:
| Dataset | sites [1500-2000] | genes [1500-2000] | total Genes | % |
| SIN3set | 8 | 7 | 22 | 31% |
| Whole genome | 288 | 137 | 13729 | 0.9% |
6. Functional annotation of 7 and 137 interesting genes:
-- 7 genes from SIN3set with the target site in 1500-2000 --
-- 137 genes from whole genome with the target site in 1500-2000 --
-- THE WHOLE GENOME --
7. Recomputing the matrices from results sets (T=0.5)
Multiple alignment of sites and matrices obtained using MEME
- SIN3 set:
- SIN3set 16 aligned sites
- SIN3set new matrix (16 sites)
Predictions with T=0.5:
- Trying again in SIN3 set (17 sites)
- Trying again in the whole genome
- Whole genome:

















