

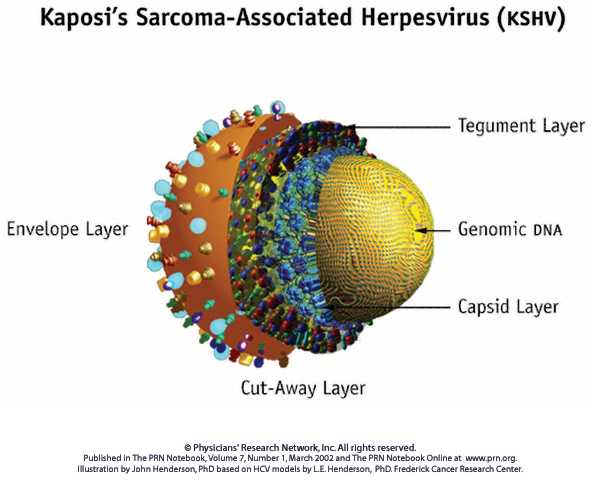
The viruses belonging to the Herpes virus Family are big and complex. Whereas they have many differences in their genome and proteins, they show many similarities in the virion structure and in their genome organization. This kind of viruses causes acute respiratory diseases that become chronic and latent , provoking many recurrent infections.
The virions of all Herpesviriae share the same structure: a core containing a linear molecule of double stranded DNA , a capsid of approximately 100-110 nm of diameter with 162 subunits (capsomers), the tegument and an envelope with viral glycoproteins in its surface. The virion contains up to 35 proteins, including some enzymes related to the nucleic acid metabolism, DNA synthesis and proteic process13.
The overall virion diameter varies from 120 to 300 nm.Variations of size depend partly on the tegument thickness and the state of the envelope, differing from one Herpes virus to another, even within the same species9.
| DNA | 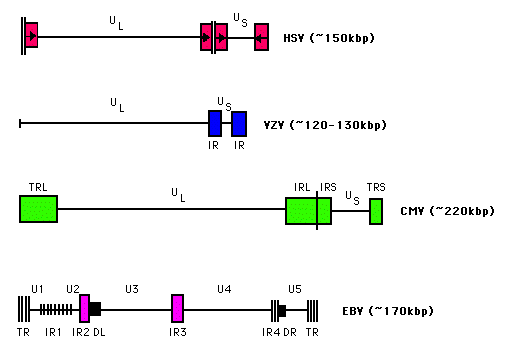 | The Herpes virus genetic material consists of a single, linear molecule of double stranded DNA which circularizes immediately after being released from capsid into the nucleous of an infected cell. DNA (figure 1), has one unique short region (US) and one unique long region (UL), flanked by many repeated sequences, inverted (IR) and/or terminal (TR). The size of the DNA is between 120 and 230 kbp, depending on the internal and terminal repeated sequences that the virion has. |
|---|---|---|
| CORE |  | The shape of the core is toroidal (Figure 2). It contains the viral DNA which in some cases is associated with a protein structure. |
| CAPSID |  | It is one of the most characteristic structures in all Herpes virus: it consists of an icosahedral capsid and its diameter is between 95 and 105 nm. It is constituted by 162 capsomers (12 pentons and 150 hexons) (Figure 3). Usually the mature capsid is composed of six different proteins. |
| TEGUMENT | 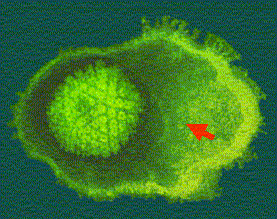 | It is a structure of amorphous material surrounding the capsid (Figure 4). It is located between de capsid and the envelope. When negatively stained has fibrous appearance. |
| ENVELOPE | 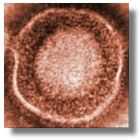 | It is a structure with a typical three-layer structure since it comes from cellular membranes partially modified and slightly pleomorphic (Figure 5). Therefore, the envelope contains lipids and numerous spicules formed by viral glycoproteins of about 8 nm size. The number of the different glycoproteins varies from one type to another. For instance, HSV-1 contains at least 11 different types. |
![]()
The equine Herpesvirus type 1 and 4
The equine Herpes virus (EHV) is the main cause of respiratory disease, abortions and neurological disorders in horses. Altough once was thought that there were two subtypes of EHV, later on it was demonstrated that in fact they were two different members of the same genus Varicellovirus, belonging to the same Subfamily Alphaherpesvirinae in the Herpesviridae Family, (Evolutionary relatedness of Herpesvirus and their Subfamilies1), they are called EHV-1 and EHV-42,3.
Only EHV-1 causes neurological disorders, while EHV-4 rarely causes abortions.

The EHV virion, like the rest of all Herpes virus (see the details of the Herpes virus structure), contains a core with a linear, doble stranded DNA, an icosahedrical capsid of 100-110 nm in diameter formed by 162 capsomers, the tegument surrounding the capsid and a lipid envelope with surface glycoproteins10.
Both genomes are collinear and have the same structure, presenting one unique long sequence (UL) joined to one unique short sequence (US) flanked by one inverted repeat (TRS IRS). In addition, they contain 12 sets of tandem repeats only three of them codify, one located in the gen 24 and the rest in the gen 712.
Another similarity is the number of gens. Both EHV-1 and EHV-4 have 76 orfs representing 77 gens, since the orfs 44 and 47 undergo splicing. Both genomes show differences in the size of their components and in their content of Cs and Ts2:
| EHV-1 | EHV-4 | |
|---|---|---|
| Genome Size | 145597 bp | 150223 bp |
| UL Size | 112871 bp | 112398 bp |
| US Size | 11861 bp | 12789 bp |
| TRS IRS | 12714 bp | 10178 bp |
| TRL IRL | 32 bp | ----- |
| Mean C/T | 56.7 % | 50.5 % |
This study is focused on the analysis of the equine Herpes virus 1 and 4 genomes at two different levels: first, prediction and further, analysis of the orfs present in their sequence.
Our objectives, therefore, are:
- Development of a computer programme (Findorf 2003, version 1.0) able to identify orfs in genomic sequences laking introns (as it is the case of the Herpes virus genomes). This implies the development of a second supporting programme (Transfasta 2003, version 1.0) with the purpose of transforming sequences to fasta format, which is the format accepted by the orfs searching programme.
- Prediction of all the possible orf in the EHV1 and EHV4 genomes using the developed programme, and further identification of those orfs already described in the literature.
- Detection of non- described orfs and investigation of their potential biological significance and function.