

1.-Abstract
For understand the transcriptional regulatory mechanism that operate in genes, it's important to realize a mapping of transcription factor binding sites in genes. Now, there are many of these site experimentally identified that can be used to perform a computational pattern-based searches. Motifs are a short and variable regions that will produce over-predictions problems.
We have developed a new program (in Perl language) for deteccion of promoter motif in a DNA sequences. Motifs,are descripted in a Position Weight Matrix (PWM) from TRANSFAC database. Position Weigth Matrix is a motif descriptor. It attempts to capture the intrinsic variability characteristic of sequence patterns. It is usually derived from a set of aligned sequences functionally related.
2.-Biological Problem
The regulation of gene expresion in eukariotes is a complex process that is difficult to understand due to variability of the mechanism involved in and the great number of different actors playing some minor o major role.
How is gene expression regulated?There are several methods used by eukaryotes.
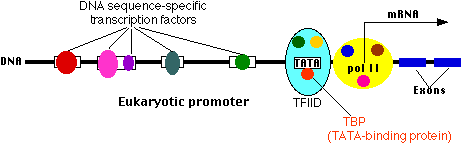
3.- The Program
Introduction
Promotif will detect promoter motif in a DNA sequence using a PWM
Inputs: Tranfac matrix and fasta sequence
ProMotif compares input PWM and input sequence. It compares every subsequence (with a position numbers like matrix- positions number)of input sequence, whit PWM, and calculate a score.
We can choose the type of PWM that ProMotif use to compute the scores:
Requests
matrix_file.mat: should be in Transfac format.
sequence_file.txt: should be in Fasta format.
Basic program ejecution
$./perl ProMotif.pl matrix_file.mat sequence_file.txt
Options
-c_x.x :Cutoff value -v :Show information of the processing -n_x :Chose type of matrix-Relative Matrix Default matrix: relative x=1 -Log-likelihood matrix(whit a priory frequency-0'25-) x=2 -Log-likelihood matrix(frequency in the aligned region) -m :Show information about the matrices -s :Show information about DNA sequences (C+G content, lenght) -o :Makes a HTML Output -h :help
Execution example:
$./perl ProMotif.pl -svc 0.55 matrix_file.mat sequence_file.txt
4.- Test
With the finality of test our program we have designed an exercise. We searched a HNF1alpha motif in a problem sequences. We obtained he HNF1alpha PWM in TRANSFAC database. To compare the results ofproblem sequences, we have maked a negative control using a aleatory sequences obtained whith a program(AleatorySequences) We run ProMotif with these input files:
| Output for problem sequences | Output for aleatory sequences | ||
| treshold | hits | treshold | hits |
| 0.95 | 1 | 0.95 | 0 |
| 0.85 | 10 | 0.85 | 0 |
| 0.80 | 33 | 0.80 | 5 |
In conclusion, we can say that ProMotif search specífic motif, and his reults aren't due to fate.
5.- Bibliography
-MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids research, 1995, Vol.23, No.23
-TRANSFAC