ORFFINDER

Authors:
Ángel Núñez
Pagán
Patricia Gordillo Blanco
Beatriz del Val Romero

____ABSTRACT____
The
aim of this program is to find ORFs (Open Reading Frames) in FASTA
format prokaryote sequences, forwards and for the 3 possible frames. Specifically,
only the ORFs located between two stop codons. So, we discard the part
of the sequence placed between the beginning and the first stop codon,
as well as the part between the last stop codon and the end of the sequence.
The program gives a score to each ORF based in a codon usage bias table.
Finally, it gives back tabulated results in an ".out" file, which can be
modified by the user.
____SPECIFICATIONS____
1)
This program is written in PERL language. So, a computer with UNIX installed
in it is needed to run it.
2)
It is very important to enter the parameters, and in the right order (that
is, first the name of the file containing the sequence, then the file containing
the codon usage bias table). Likewise, it has to be checked that both the
sequence and the table are written in the DNA code.
3)
It is necessary to check that the sequence and table formats are the right
ones. At the moment of downloading the table at the web page recommended
it is advisable to follow the next steps:
- select the organism
- submit the obtained table, selecting "standard" format and "A style like
Codon Frequency output in
GCG"
- click on "submit" and save the outcoming table at a .txt file
- run TRANSFORMER with the .txt file to get the same table in the right
format to run ORFFINDER
4)
Once obtained the ORFFINDER outfile, it can be filtered to obtain, in other
outfiles, the ORFs that are more probable to be coding. The cut-off is
fixed by the user through UNIX commands. It can be applied not to only
to the score, but also to the length of the ORFs.
5)
The filtered file has to be turned into GFF format to view the results
graphically. It can be done using a UNIX command.
6)
The GFF file transforms into PS (Postscript) file through the GFF2PS program
.
7)
The GHOSTVIEW program, which can be found in SHELL, is used to view the
PS file at screen.
NOTE: all the necessary
UNIX commands are illustrated below.
____LINKS___
____EXAMPLE____
-
We
start from SHELL and run ORFFINDER with the following statement:
./orffinder.pl secuencia.fa tabla.txt
The name of the files is illustrative, it can be chosen.
The sequence (first parameter) entry in NCBI
can be found here.
The usage codon table (second parameter) comes from the web page suggested
above.
-
The
program will run and turn us back a file called "resultados.out"
(results.out).
-
We filter the results' file
with the UNIX "gawk" command. In our case, we have decided to apply a double
filter to remove those ORFs with a size ($4) lower than 200 bases and negative
score ($5). The result of the filtration is directed to "resultados_filtro.out"
(results_filter.out):
gawk '($1 ~ /[012]/ && $4>200 && $5>0) { print $0 }' resultados.out
> resultados_filtro.out
NOTE: The parameters of the filter are changeable.
-
We
transform the .out file formerly generated to a GFF format file through
the following UNIX command, obtaining a file called "resultados_filtro.gff"
(results_filter.gff):
gawk 'BEGIN{ OFS="\t" } $1 ~ /[012]/ { print $1, "orffinder", "orf",
$2, $3, $5, $6, 0 }' resultados_filtro.out > resultados_filtro.gff
-
Now
it is necessary to turn the GFF file into a PS file, which we can view
by GHOSTVIEW. In order to make, we use the program GFF2PS (you can download
it from the link placed in the previous section). The command to run
GFF2PS with the default parameters is the following. We redirect the results
to the file "resultados_filtro.ps"
(results_filter.ps).
./gff2ps -Vr -- resultados_filtro.gff > resultados_filtro.ps
NOTE:
To modify the format of the .ps file obtained, several GFF2PS parameters
can be used. They can be found in the user's manual in the GFF2PS web page.
-
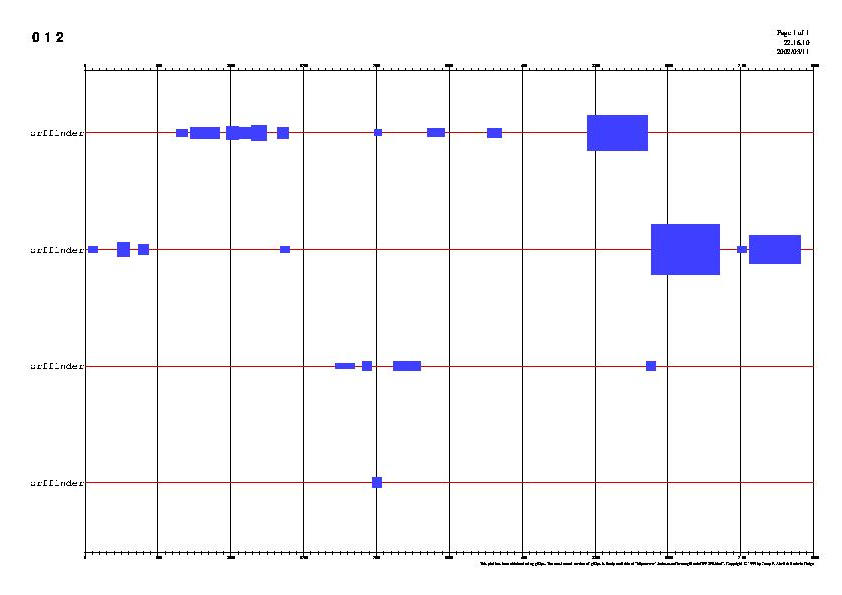
To
view the PS file we open it with the GHOSTVIEW program from SHELL, using
the following command:
./ghostview -landscape resultados_filtro.ps
-
The
PS file can be turned into a JPEG one with the next command (optional):
ghostscript -dBATCH -dNOPAUSE -sPAPERSIZE=a4 -sDEVICE=jpeg -sOutputFile=resultados_filtro.jpg
resultados_filtro.ps
-
It
is also optional to rotate the image to make easier the interpretation
of the results:
convert -rotate 90 resultados_filtro.jpg resultados_filtro_rot.jpg
ACKNOWLEDGEMENTS
To
all the people that helped us (¡ya sabeis quien sois!)





